Genomics analysis of Drosophila sechellia response to Morinda citrifolia fruit diet
- PMID: 35736356
- PMCID: PMC9526069
- DOI: 10.1093/g3journal/jkac153
Genomics analysis of Drosophila sechellia response to Morinda citrifolia fruit diet
Abstract
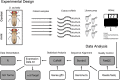
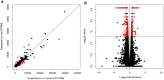
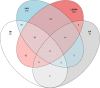
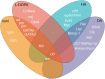
Drosophila sechellia is an island endemic host specialist that has evolved to consume the toxic fruit of Morinda citrifolia, also known as noni fruit. Recent studies by our group and others have examined genome-wide gene expression responses of fruit flies to individual highly abundant compounds found in noni responsible for the fruit's unique chemistry and toxicity. In order to relate these reductionist experiments to the gene expression responses to feeding on noni fruit itself, we fed rotten noni fruit to adult female D. sechellia and performed RNA-sequencing. Combining the reductionist and more wholistic approaches, we have identified candidate genes that may contribute to each individual compound and those that play a more general role in response to the fruit as a whole. Using the compound specific and general responses, we used transcription factor prediction analyses to identify the regulatory networks and specific regulators involved in the responses to each compound and the fruit itself. The identified genes and regulators represent the possible genetic mechanisms and biochemical pathways that contribute to toxin resistance and noni specialization in D. sechellia.
Keywords: Morinda citrifolia; RNA-seq; host specialization; noni; toxin resistance.
© The Author(s) 2022. Published by Oxford University Press on behalf of Genetics Society of America.
Conflict of interest statement
None declared.
Figures

References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. Available at: http://Www.Bioinformatics.Babraham.Ac.Uk/Projects/Fastqc.
-
- Benjamini Y, Hochberg Y. Controlling the false discovery rate : a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 1995;57(1):289–300.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
