A Novel Probiotic Bacillus subtilis Strain Confers Cytoprotection to Host Pig Intestinal Epithelial Cells during Enterotoxic Escherichia coli Infection
- PMID: 35736372
- PMCID: PMC9430607
- DOI: 10.1128/spectrum.01257-21
A Novel Probiotic Bacillus subtilis Strain Confers Cytoprotection to Host Pig Intestinal Epithelial Cells during Enterotoxic Escherichia coli Infection
Abstract
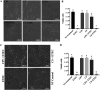
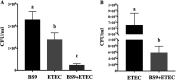
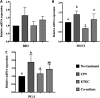
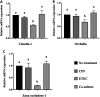
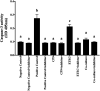
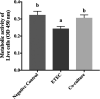
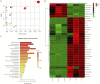
Enteric infections caused by enterotoxic Escherichia coli (ETEC) negatively impact the growth performance of piglets during weaning, resulting in significant economic losses for the producers. With the ban on antibiotic usage in livestock production, probiotics have gained a lot of attention as a potential alternative. However, strain specificity and limited knowledge on the host-specific targets limit their efficacy in preventing ETEC-related postweaning enteric infections. We recently isolated and characterized a novel probiotic Bacillus subtilis bacterium (CP9) that demonstrated antimicrobial activity. Here, we report anti-ETEC properties of CP9 and its impact on metabolic activity of swine intestinal epithelial (IPEC-J2) cells. Our results showed that pre- or coincubation with CP9 protected IPEC-J2 cells from ETEC-induced cytotoxicity. CP9 significantly attenuated ETEC-induced inflammatory response by reducing ETEC-induced nitric oxide production and relative mRNA expression of the Toll-like receptors (TLRs; TLR2, TLR4, and TLR9), proinflammatory tumor necrosis factor alpha, interleukins (ILs; IL-6 and IL-8), augmenting anti-inflammatory granulocyte-macrophage colony-stimulating factor and host defense peptide mucin 1 (MUC1) mRNA levels. We also show that CP9 significantly (P < 0.05) reduced caspase-3 activity, reinstated cell proliferation and increased relative expression of tight junction genes, claudin-1, occludin, and zona occludens-1 in ETEC-infected cells. Finally, metabolomic analysis revealed that CP9 exposure induced metabolic modulation in IPEC J2 cells with the greatest impact seen in alanine, aspartate, and glutamate metabolism; pyrimidine metabolism; nicotinate and nicotinamide metabolism; glutathione metabolism; the citrate cycle (TCA cycle); and arginine and proline metabolism. Our study shows that CP9 incubation attenuated ETEC-induced cytotoxicity in IPEC-J2 cells and offers insight into potential application of this probiotic for ETEC infection control. IMPORTANCE ETEC remains one of the leading causes of postweaning diarrhea and mortality in swine production. Due to the rising concerns with the antibiotic use in livestock, alternative interventions need to be developed. In this study, we analyzed the cytoprotective effect of a novel probiotic strain in combating ETEC infection in swine intestinal cells, along with assessing its mechanism of action. To our knowledge, this is also the first study to analyze the metabolic impact of a probiotic on intestinal cells. Results from this study should provide effective cues in developing a probiotic intervention for ameliorating ETEC infection and improving overall gut health in swine production.
Keywords: Bacillus subtilis; antimicrobial; antimicrobial agents; cytoprotection; enterotoxic E. coli; extreme environment; probiotic.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Bednorz C, Oelgeschläger K, Kinnemann B, Hartmann S, Neumann K, Pieper R, Bethe A, Semmler T, Tedin K, Schierack P, Wieler LH, Guenther S. 2013. The broader context of antibiotic resistance: zinc feed supplementation of piglets increases the proportion of multi-resistant Escherichia coli in vivo. Int J Med Microbiol 303:396–403. doi:10.1016/j.ijmm.2013.06.004. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

