Association between Triplex-Forming Sites of Cardiac Long Noncoding RNA GATA6-AS1 and Chromatin Organization
- PMID: 35736638
- PMCID: PMC9227037
- DOI: 10.3390/ncrna8030041
Association between Triplex-Forming Sites of Cardiac Long Noncoding RNA GATA6-AS1 and Chromatin Organization
Abstract
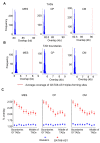
This study explored the relationship between 3D genome organization and RNA-DNA triplex-forming sites of long noncoding RNAs (lncRNAs), a group of RNAs that do not code for proteins but are important factors regulating different aspects of genome activity. The triplex-forming sites of anti-sense cardiac lncRNA GATA6-AS1 derived from DBD-Capture-Seq were examined and compared to modular features of 3D genome organization called topologically associated domains (TADs) obtained from Hi-C data. It was found that GATA6-AS1 triplex-forming sites are positioned non-randomly in TADs and their boundaries. The triplex sites showed a preference for TAD boundaries over internal regions of TADs. Computational prediction analysis indicated that CTCF, the key protein involved in TAD specification, may interact with GATA6-AS1, and their binding sites correlate with each other. Examining locations of repeat elements in the genome suggests that the ability of lncRNA GATA6-AS1 to form triplex sites with many genomic locations may be achieved by the rapid expansion of different repeat elements. Some of the triplex-forming sites were found to be positioned in regions that undergo dynamic chromatin organization events such as loss/gain of TAD boundaries during cardiac differentiation. These observed associations suggest that lncRNA-DNA triplex formation may contribute to the specification of TADs in 3D genome organization.
Keywords: CTCF; GATA6-AS1; RNA–DNA triplex; topologically associated domains.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources

