A Comprehensive Study of the WRKY Transcription Factor Family in Strawberry
- PMID: 35736736
- PMCID: PMC9229891
- DOI: 10.3390/plants11121585
A Comprehensive Study of the WRKY Transcription Factor Family in Strawberry
Abstract
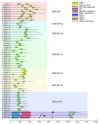
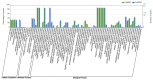
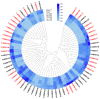
WRKY transcription factors play critical roles in plant growth and development or stress responses. Using up-to-date genomic data, a total of 64 and 257 WRKY genes have been identified in the diploid woodland strawberry, Fragaria vesca, and the more complex allo-octoploid commercial strawberry, Fragaria × ananassa cv. Camarosa, respectively. The completeness of the new genomes and annotations has enabled us to perform a more detailed evolutionary and functional study of the strawberry WRKY family members, particularly in the case of the cultivated hybrid, in which homoeologous and paralogous FaWRKY genes have been characterized. Analysis of the available expression profiles has revealed that many strawberry WRKY genes show preferential or tissue-specific expression. Furthermore, significant differential expression of several FaWRKY genes has been clearly detected in fruit receptacles and achenes during the ripening process and pathogen challenged, supporting a precise functional role of these strawberry genes in such processes. Further, an extensive analysis of predicted development, stress and hormone-responsive cis-acting elements in the strawberry WRKY family is shown. Our results provide a deeper and more comprehensive knowledge of the WRKY gene family in strawberry.
Keywords: WRKY family; differential expression; homeolog; paralog; pathogens; ripening; strawberry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources

