Unraveling NPR-like Family Genes in Fragaria spp. Facilitated to Identify Putative NPR1 and NPR3/4 Orthologues Participating in Strawberry- Colletotrichum fructicola Interaction
- PMID: 35736739
- PMCID: PMC9229442
- DOI: 10.3390/plants11121589
Unraveling NPR-like Family Genes in Fragaria spp. Facilitated to Identify Putative NPR1 and NPR3/4 Orthologues Participating in Strawberry- Colletotrichum fructicola Interaction
Abstract
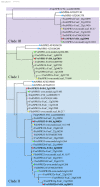
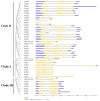
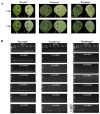
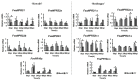
The salicylic acid receptor NPR1 (nonexpressor of pathogenesis-related genes) and its paralogues NPR3 and NPR4 are master regulators of plant immunity. Commercial strawberry (Fragaria × ananassa) is a highly valued crop vulnerable to various pathogens. Historic confusions regarding the identity of NPR-like genes have hindered research in strawberry resistance. In this study, the comprehensive identification and phylogenic analysis unraveled this family, harboring 6, 6, 5, and 23 members in F. vesca, F. viridis, F. iinumae, and F. × ananassa, respectively. These genes were clustered into three clades, with each diploid member matching three to five homoalleles in F. × ananassa. Despite the high conservation in terms of gene structure, protein module, and functional residues/motifs/domains, substantial divergence was observed, hinting strawberry NPR proteins probably function in ways somewhat different from Arabidopsis. RT-PCR and RNAseq analysis evidenced the transcriptional responses of FveNPR1 and FxaNPR1a to Colletotrichum fructicola. Extended expression analysis for strawberry NPR-likes helped to us understand how strawberry orchestrate the NPRs-centered defense system against C. fructicola. The cThe current work supports that FveNPR1 and FxaNPR1a, as well as FveNPR31 and FxaNPR31a-c, were putative functional orthologues of AtNPR1 and AtNPR3/4, respectively. These findings set a solid basis for the molecular dissection of biological functions of strawberry NPR-like genes for improving disease resistance.
Keywords: NPR-like family; anthracnose; expression profile; phylogeny; strawberry.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






References
-
- Saleh A., Withers J., Mohan R., Marqués J., Gu Y., Yan S., Zavaliev R., Nomoto M., Tada Y., Dong X. Posttranslational Modifications of the Master Transcriptional Regulator NPR1 Enable Dynamic but Tight Control of Plant Immune Responses. Cell Host Microbe. 2015;18:169–182. doi: 10.1016/j.chom.2015.07.005. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

