A Comparison of Blood Plasma Small Extracellular Vesicle Enrichment Strategies for Proteomic Analysis
- PMID: 35736799
- PMCID: PMC9229025
- DOI: 10.3390/proteomes10020019
A Comparison of Blood Plasma Small Extracellular Vesicle Enrichment Strategies for Proteomic Analysis
Abstract
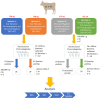
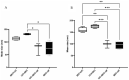
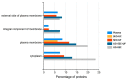
Proteomic analysis of small extracellular vesicles (sEVs) poses a significant challenge. A 'gold-standard' method for plasma sEV enrichment for downstream proteomic analysis is yet to be established. Methods were evaluated for their capacity to successfully isolate and enrich sEVs from plasma, minimise the presence of highly abundant plasma proteins, and result in the optimum representation of sEV proteins by liquid chromatography tandem mass spectrometry. Plasma from four cattle (Bos taurus) of similar physical attributes and genetics were used. Three methods of sEV enrichment were utilised: ultracentrifugation (UC), size-exclusion chromatography (SEC), and ultrafiltration (UF). These methods were combined to create four groups for methodological evaluation: UC + SEC, UC + SEC + UF, SEC + UC and SEC + UF. The UC + SEC method yielded the highest number of protein identifications (IDs). The SEC + UC method reduced plasma protein IDs compared to the other methods, but also resulted in the lowest number of protein IDs overall. The UC + SEC + UF method decreased sEV protein ID, particle number, mean and mode particle size, particle yield, and did not improve purity compared to the UC + SEC method. In this study, the UC + SEC method was the best method for sEV protein ID, purity, and overall particle yield. Our data suggest that the method and sequence of sEV enrichment strategy impacts protein ID, which may influence the outcome of biomarker discovery studies.
Keywords: enrichment; exosome; extracellular vesicle; extracellular vesicles; isolation; mass spectrometry; proteomics; size-exclusion chromatography; small extracellular vesicle; ultracentrifugation; ultrafiltration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Collins B.C., Hunter C.L., Liu Y., Schilling B., Rosenberger G., Bader S.L., Chan D.W., Gibson B.W., Gingras A.-C., Held J.M., et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 2017;8:291. doi: 10.1038/s41467-017-00249-5. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

