Glycan degradation promotes macroautophagy
- PMID: 35737835
- PMCID: PMC9245654
- DOI: 10.1073/pnas.2111506119
Glycan degradation promotes macroautophagy
Abstract
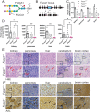
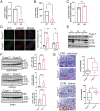
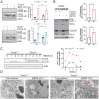
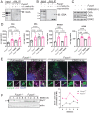
Macroautophagy promotes cellular homeostasis by delivering cytoplasmic constituents to lysosomes for degradation [Mizushima, Nat. Cell Biol. 20, 521-527 (2018)]. However, while most studies have focused on the mechanisms of protein degradation during this process, we report here that macroautophagy also depends on glycan degradation via the glycosidase, α-l-fucosidase 1 (FUCA1), which removes fucose from glycans. We show that cells lacking FUCA1 accumulate lysosomal glycans, which is associated with impaired autophagic flux. Moreover, in a mouse model of fucosidosis-a disease characterized by inactivating mutations in FUCA1 [Stepien et al., Genes (Basel) 11, E1383 (2020)]-glycan and autophagosome/autolysosome accumulation accompanies tissue destruction. Mechanistically, using lectin capture and mass spectrometry, we identified several lysosomal enzymes with altered fucosylation in FUCA1-null cells. Moreover, we show that the activity of some of these enzymes in the absence of FUCA1 can no longer be induced upon autophagy stimulation, causing retardation of autophagic flux, which involves impaired autophagosome-lysosome fusion. These findings therefore show that dysregulated glycan degradation leads to defective autophagy, which is likely a contributing factor in the etiology of fucosidosis.
Keywords: fucosidosis; lysosomes; macroautophagy; α-l-fucosidase 1.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

