Molecular profiling identifies targeted therapy opportunities in pediatric solid cancer
- PMID: 35739269
- PMCID: PMC10953704
- DOI: 10.1038/s41591-022-01856-6
Molecular profiling identifies targeted therapy opportunities in pediatric solid cancer
Abstract
To evaluate the clinical impact of molecular tumor profiling (MTP) with targeted sequencing panel tests, pediatric patients with extracranial solid tumors were enrolled in a prospective observational cohort study at 12 institutions. In the 345-patient analytical population, median age at diagnosis was 12 years (range 0-27.5); 298 patients (86%) had 1 or more alterations with potential for impact on care. Genomic alterations with diagnostic, prognostic or therapeutic significance were present in 61, 16 and 65% of patients, respectively. After return of the results, impact on care included 17 patients with a clarified diagnostic classification and 240 patients with an MTP result that could be used to select molecularly targeted therapy matched to identified alterations (MTT). Of the 29 patients who received MTT, 24% had an objective response or experienced durable clinical benefit; all but 1 of these patients received targeted therapy matched to a gene fusion. Of the diagnostic variants identified in 209 patients, 77% were gene fusions. MTP with targeted panel tests that includes fusion detection has a substantial clinical impact for young patients with solid tumors.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
A.J.C. sits on an advisory board for Bayer. L.B.C. is an employee at Sema4 and is a consultant for X-Chem and Biomatics. L.M. is a consultant for Jazz Pharmaceuticals. T.W.L. is an advisory board member for Bayer and has received honoraria from Bayer, Cellectis, Novartis, Deciphera, Jumo Health and Y-mAbs and has received research funding from Pfizer and Bayer. M.A.A. is an advisory board member for Fennec Pharmaceuticals. Y-C.L. is an advisory board member for Takeda Pharmaceutical Company. A.D.C. has received research support from Bayer and his spouse is employed by Labcorp. J.K.’s spouse has received consulting fees from ROME Therapeutics, Foundation Medicine, NanoString, Merck Millipore and Pfizer that are not related to this work. J.K.’s spouse is a founder and has equity in ROME Therapeutics, PanTher Therapeutics and TellBio, which is not related to this work. J.K.’s spouse receives research support from ACD Bio-Techne, PureTech Health and Ribon Therapeutics, which was not used in this work. S.L.V. is a consultant for CVS Accordant. E.V.A. has provided advisory/consulting work for Tango Therapeutics, Genome Medical, Invitae, Enara Bio, Janssen, Manifold Bio and Monte Rosa Therapeutics, has received research support from Novartis and Bristol Myers Squibb, holds equity in Tango Therapeutics, Genome Medical, Syapse, Enara Bio, Manifold Bio, Microsoft and Monte Rosa Therapeutics, has received travel reimbursement from Roche/Genentech and holds institutional patents filed on chromatin mutations and immunotherapy response and methods for clinical interpretation. S.G.D. has consulted for Bayer and received travel expenses from Loxo Oncology, Roche and Salarius. W.B.L. has served on the data safety monitoring boards for Merck Millipore and Jubilant Draximage. K.A.J. has consulted for Ipsen and Bayer, and has received honoraria from Foundation Medicine and Takeda Pharmaceutical Company. The other authors declare no competing interests.
Figures
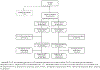









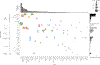



Comment in
-
Pediatric precision oncology-Time for clinical prime time.Med. 2022 Aug 12;3(8):521-522. doi: 10.1016/j.medj.2022.07.004. Med. 2022. PMID: 35963231
References
-
- Lindeman NI et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J. Thorac. Oncol 13, 323–358 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

