Single-cell transcriptomics and surface epitope detection in human brain epileptic lesions identifies pro-inflammatory signaling
- PMID: 35739273
- PMCID: PMC9276529
- DOI: 10.1038/s41593-022-01095-5
Single-cell transcriptomics and surface epitope detection in human brain epileptic lesions identifies pro-inflammatory signaling
Abstract
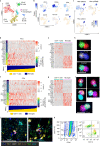
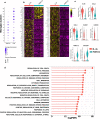
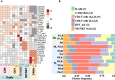
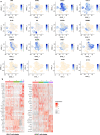
Epileptogenic triggers are multifactorial and not well understood. Here we aimed to address the hypothesis that inappropriate pro-inflammatory mechanisms contribute to the pathogenesis of refractory epilepsy (non-responsiveness to antiepileptic drugs) in human patients. We used single-cell cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) to reveal the immunotranscriptome of surgically resected epileptic lesion tissues. Our approach uncovered a pro-inflammatory microenvironment, including extensive activation of microglia and infiltration of other pro-inflammatory immune cells. These findings were supported by ligand-receptor (LR) interactome analysis, which demonstrated potential mechanisms of infiltration and evidence of direct physical interactions between microglia and T cells. Together, these data provide insight into the immune microenvironment in epileptic tissue, which may aid the development of new therapeutics.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











Comment in
-
Neuro-immune crosstalk in drug-resistant epilepsy.Nat Neurosci. 2022 Jul;25(7):843-844. doi: 10.1038/s41593-022-01112-7. Nat Neurosci. 2022. PMID: 35739272 No abstract available.
-
Exploring CITEs of Inflammation in the Human Epilepsy Brain.Epilepsy Curr. 2023 Apr 11;23(3):191-192. doi: 10.1177/15357597231161460. eCollection 2023 May-Jun. Epilepsy Curr. 2023. PMID: 37334410 Free PMC article. No abstract available.
References
-
- Reynolds EH. The ILAE/IBE/WHO global campaign against epilepsy: bringing epilepsy “out of the shadows”. Epilepsy Behav. 2000;1:S3–S8. - PubMed
-
- Epilepsy: Key Facts. WHOhttps://www.who.int/ (2018).
-
- Lerche H. Drug-resistant epilepsy—time to target mechanisms. Nat. Rev. Neurol. 2020;16:595–596. - PubMed
-
- Ravizza T, et al. Innate and adaptive immunity during epileptogenesis and spontaneous seizures: evidence from experimental models and human temporal lobe epilepsy. Neurobiol. Dis. 2008;29:142–160. - PubMed
-
- Auvin S, Cilio MR, Vezzani A. Current understanding and neurobiology of epileptic encephalopathies. Neurobiol. Dis. 2016;92:72–89. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

