Identification of Genomic Regions and Candidate Genes for Litter Traits in French Large White Pigs Using Genome-Wide Association Studies
- PMID: 35739920
- PMCID: PMC9219640
- DOI: 10.3390/ani12121584
Identification of Genomic Regions and Candidate Genes for Litter Traits in French Large White Pigs Using Genome-Wide Association Studies
Abstract
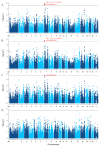
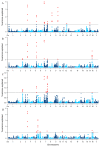
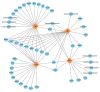
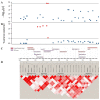
The reproductive traits of sows are one of the important economic traits in pig production, and their performance directly affects the economic benefits of the entire pig industry. In this study, a total of 895 French Large White pigs were genotyped by GeneSeek Porcine 50K SNP Beadchip and four phenotypic traits of 1407 pigs were recorded, including total number born (TNB), number born alive (NBA), number healthy piglets (NHP) and litter weight born alive (LWB). To identify genomic regions and genes for these traits, we used two approaches: a single-locus genome-wide association study (GWAS) and a single-step GWAS (ssGWAS). Overall, a total of five SNPs and 36 genomic regions were identified by single-locus GWAS and ssGWAS, respectively. Notably, fourof all five significant SNPs were located in 10.72-11.06 Mb on chromosome 7, were also identified by ssGWAS. These regions explained the highest or second highest genetic variance in the TNB, NBA and NHP traits and harbor the protein coding gene ENSSSCG00000042180. In addition, several candidate genes associated with litter traits were identified, including JARID2, PDIA6, FLRT2 and DICER1. Overall, these novel results reflect the polygenic genetic architecture of the litter traits and provide a theoretical reference for the following implementation of molecular breeding.
Keywords: SNP; SsGWAS; genetic variants; pig; reproductive traits.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

