Bioenergetic and Autophagic Characterization of Skin Fibroblasts from C9orf72 Patients
- PMID: 35740026
- PMCID: PMC9219955
- DOI: 10.3390/antiox11061129
Bioenergetic and Autophagic Characterization of Skin Fibroblasts from C9orf72 Patients
Abstract
The objective of this study is to describe the alterations occurring during the neurodegenerative process in skin fibroblast cultures from C9orf72 patients. We characterized the oxidative stress, autophagy flux, small ubiquitin-related protein SUMO2/3 levels as well as the mitochondrial function in skin fibroblast cultures from C9orf72 patients. All metabolic and bioenergetic findings were further correlated with gene expression data obtained from RNA sequencing analysis. Fibroblasts from C9orf72 patients showed a 30% reduced expression of C9orf72, ~3-fold increased levels of oxidative stress and impaired mitochondrial function obtained by measuring the enzymatic activities of mitochondrial respiratory chain complexes, specifically of complex III activity. Furthermore, the results also reveal that C9orf72 patients showed an accumulation of p62 protein levels, suggesting the alteration of the autophagy process, and significantly higher protein levels of SUMO2/3 (p = 0.03). Our results provide new data reinforcing that C9orf72 cells suffer from elevated oxidative damage to biomolecules and organelles and from increased protein loads, leading to insufficient autophagy and an increase in SUMOylation processes.
Keywords: C9orf72; SUMOlyation; autophagy; bioenergetics.
Conflict of interest statement
R.S.-V. has served at scientific advisory boards from Ionnis Pharmaceuticals and Wave Life Science. The other authors declare no conflicts of interest.
Figures



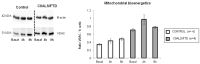

Similar articles
-
FTLD Patient-Derived Fibroblasts Show Defective Mitochondrial Function and Accumulation of p62.Mol Neurobiol. 2021 Nov;58(11):5438-5458. doi: 10.1007/s12035-021-02475-x. Epub 2021 Jul 30. Mol Neurobiol. 2021. PMID: 34328616 Free PMC article.
-
Mitochondrial and autophagic alterations in skin fibroblasts from Parkinson disease patients with Parkin mutations.Aging (Albany NY). 2019 Jun 9;11(11):3750-3767. doi: 10.18632/aging.102014. Aging (Albany NY). 2019. PMID: 31180333 Free PMC article.
-
C9orf72 ALS-FTD: recent evidence for dysregulation of the autophagy-lysosome pathway at multiple levels.Autophagy. 2021 Nov;17(11):3306-3322. doi: 10.1080/15548627.2021.1872189. Epub 2021 Feb 26. Autophagy. 2021. PMID: 33632058 Free PMC article. Review.
-
Systemic deregulation of autophagy upon loss of ALS- and FTD-linked C9orf72.Autophagy. 2017 Jul 3;13(7):1254-1255. doi: 10.1080/15548627.2017.1299312. Autophagy. 2017. PMID: 28319438 Free PMC article.
-
Bioenergetics and Autophagic Imbalance in Patients-Derived Cell Models of Parkinson Disease Supports Systemic Dysfunction in Neurodegeneration.Front Neurosci. 2019 Sep 10;13:894. doi: 10.3389/fnins.2019.00894. eCollection 2019. Front Neurosci. 2019. PMID: 31551675 Free PMC article. Review.
Cited by
-
C9orf72-Associated Dipeptide Repeat Expansions Perturb ER-Golgi Vesicular Trafficking, Inducing Golgi Fragmentation and ER Stress, in ALS/FTD.Mol Neurobiol. 2024 Dec;61(12):10318-10338. doi: 10.1007/s12035-024-04187-4. Epub 2024 May 9. Mol Neurobiol. 2024. PMID: 38722513 Free PMC article.
-
Molecular mechanism of vimentin nuclear localization associated with the migration and invasion of daughter cells derived from polyploid giant cancer cells.J Transl Med. 2023 Oct 13;21(1):719. doi: 10.1186/s12967-023-04585-7. J Transl Med. 2023. PMID: 37833712 Free PMC article.
-
VDAC1: A Key Player in the Mitochondrial Landscape of Neurodegeneration.Biomolecules. 2024 Dec 30;15(1):33. doi: 10.3390/biom15010033. Biomolecules. 2024. PMID: 39858428 Free PMC article. Review.
-
Altered TDP-43 Structure and Function: Key Insights into Aberrant RNA, Mitochondrial, and Cellular and Systemic Metabolism in Amyotrophic Lateral Sclerosis.Metabolites. 2022 Jul 29;12(8):709. doi: 10.3390/metabo12080709. Metabolites. 2022. PMID: 36005581 Free PMC article. Review.
References
-
- DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. - DOI - PMC - PubMed
-
- Renton A.E., Majounie E., Waite A., Simon-Saánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. - DOI - PMC - PubMed
-
- Lee Y.-B., Chen H.-J., Peres J.N., Gomez-Deza J., Attig J., Štalekar M., Troakes C., Nishimura A.L., Scotter E.L., Vance C., et al. Hexanucleotide Repeats in ALS/FTD Form Length-Dependent RNA Foci, Sequester RNA Binding Proteins, and Are Neurotoxic. Cell Rep. 2013;5:1178–1186. doi: 10.1016/j.celrep.2013.10.049. - DOI - PMC - PubMed

