A Mutation in Endogenous saRNA miR-23a Influences Granulosa Cells Response to Oxidative Stress
- PMID: 35740072
- PMCID: PMC9219974
- DOI: 10.3390/antiox11061174
A Mutation in Endogenous saRNA miR-23a Influences Granulosa Cells Response to Oxidative Stress
Abstract
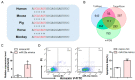
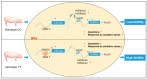
Phenotypes are the result of the interaction between the gene and the environment, so the response of individuals with different genotypes to an environment is variable. Here, we reported that a mutation in miR-23a influences granulosa cells (GCs) response to oxidative stress, a common mechanism of environmental factors affecting female reproduction. We showed that nuclear miR-23a is a pro-apoptotic miRNA in porcine GCs through the activation of the transcription and function of NORHA, a long non-coding RNA (lncRNA) induces GC apoptosis and responses to oxidative stress. Mechanistically, miR-23a acts as an endogenous small activating RNA (saRNA) to alter histone modifications of the NORHA promoter through the direct binding to its core promoter. A C > T mutation was identified at −398 nt of the miR-23a core promoter, which created a novel binding site for the transcription factor SMAD4 and recruited the transcription repressor SMAD4 to inhibit miR-23a transcription and function in GCs. Notably, g.−398C > T mutation in the miR-23a promoter reduced GCs response to oxidative stress. In addition, g.−398C > T mutation was significantly associated with sow fertility traits. In short, our findings preliminarily revealed the genetic basis of individual differences in the response to oxidative stress from the perspective of a single mutation and identified miR-23a as a candidate gene for the environmental adaptation to oxidative stress.
Keywords: GC apoptosis; NORHA-FoxO1 pathway; miR-23a; mutation; oxidative stress; saRNA.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures









References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

