Bacterial Resistance to Antibiotics and Clonal Spread in COVID-19-Positive Patients on a Tertiary Hospital Intensive Care Unit, Czech Republic
- PMID: 35740188
- PMCID: PMC9219711
- DOI: 10.3390/antibiotics11060783
Bacterial Resistance to Antibiotics and Clonal Spread in COVID-19-Positive Patients on a Tertiary Hospital Intensive Care Unit, Czech Republic
Abstract
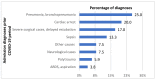
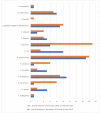
This observational retrospective study aimed to analyze whether/how the spectrum of bacterial pathogens and their resistance to antibiotics changed during the worst part of the COVID-19 pandemic (1 November 2020 to 30 April 2021) among intensive care patients in University Hospital Olomouc, Czech Republic, as compared with the pre-pandemic period (1 November 2018 to 30 April 2019). A total of 789 clinically important bacterial isolates from 189 patients were cultured during the pre-COVID-19 period. The most frequent etiologic agents causing nosocomial infections were strains of Klebsiella pneumoniae (17%), Pseudomonas aeruginosa (11%), Escherichia coli (10%), coagulase-negative staphylococci (9%), Burkholderia multivorans (8%), Enterococcus faecium (6%), Enterococcus faecalis (5%), Proteus mirabilis (5%) and Staphylococcus aureus (5%). Over the comparable COVID-19 period, a total of 1500 bacterial isolates from 372 SARS-CoV-2-positive patients were assessed. While the percentage of etiological agents causing nosocomial infections increased in Enterococcus faecium (from 6% to 19%, p < 0.0001), Klebsiella variicola (from 1% to 6%, p = 0.0004) and Serratia marcescens (from 1% to 8%, p < 0.0001), there were significant decreases in Escherichia coli (from 10% to 3%, p < 0.0001), Proteus mirabilis (from 5% to 2%, p = 0.004) and Staphylococcus aureus (from 5% to 2%, p = 0.004). The study demonstrated that the changes in bacterial resistance to antibiotics are ambiguous. An increase in the frequency of ESBL-positive strains of some species (Serratia marcescens and Enterobacter cloacae) was confirmed; on the other hand, resistance decreased (Escherichia coli, Acinetobacter baumannii) or the proportion of resistant strains remained unchanged over both periods (Klebsiella pneumoniae, Enterococcus faecium). Changes in pathogen distribution and resistance were caused partly due to antibiotic selection pressure (cefotaxime consumption increased significantly in the COVID-19 period), but mainly due to clonal spread of identical bacterial isolates from patient to patient, which was confirmed by the pulse field gel electrophoresis methodology. In addition to the above shown results, the importance of infection prevention and control in healthcare facilities is discussed, not only for dealing with SARS-CoV-2 but also for limiting the spread of bacteria.
Keywords: COVID-19; antibiotics; bacteria; multidrug resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Situation by Region, Country, Territory & Area—WHO Coronavirus (COVID-19) Dashboard. [(accessed on 30 May 2022)]. Available online: https://covid19.who.int/table.
-
- O’Neill J. Review on Antimicrobial Resistance Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. The Review on Antimicrobial Resistance; London, UK: 2014. [(accessed on 20 July 2021)]. Available online: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Ta....
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

