Unveiling the Native Morphology of Extracellular Vesicles from Human Cerebrospinal Fluid by Atomic Force and Cryogenic Electron Microscopy
- PMID: 35740275
- PMCID: PMC9220600
- DOI: 10.3390/biomedicines10061251
Unveiling the Native Morphology of Extracellular Vesicles from Human Cerebrospinal Fluid by Atomic Force and Cryogenic Electron Microscopy
Abstract
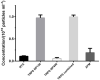
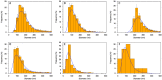
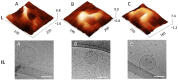
Extracellular vesicles (EVs) are membranous structures in biofluids with enormous diagnostic/prognostic potential for application in liquid biopsies. Any such downstream application requires a detailed characterization of EV concentration, size and morphology. This study aimed to observe the native morphology of EVs in human cerebrospinal fluid after traumatic brain injury. Therefore, they were separated by gravity-driven size-exclusion chromatography (SEC) and investigated by atomic force microscopy (AFM) in liquid and cryogenic transmission electron microscopy (cryo-TEM). The enrichment of EVs in early SEC fractions was confirmed by immunoblot for transmembrane proteins CD9 and CD81. These fractions were then pooled, and the concentration and particle size distribution were determined by Tunable Resistive Pulse Sensing (around 1010 particles/mL, mode 100 nm) and Nanoparticle Tracking Analysis (around 109 particles/mL, mode 150 nm). Liquid AFM and cryo-TEM investigations showed mode sizes of about 60 and 90 nm, respectively, and various morphology features. AFM revealed round, concave, multilobed EV structures; and cryo-TEM identified single, double and multi-membrane EVs. By combining AFM for the surface morphology investigation and cryo-TEM for internal structure differentiation, EV morphological subpopulations in cerebrospinal fluid could be identified. These subpopulations should be further investigated because they could have different biological functions.
Keywords: atomic force microscopy; cerebrospinal fluid; cryogenic transmission electron microscopy; extracellular vesicles; morphology; size-exclusion chromatography.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. Proteomic Comparison Defines Novel Markers to Characterize Heterogeneous Populations of Extracellular Vesicle Subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. - DOI - PMC - PubMed
-
- Li M., Huang L., Chen J., Ni F., Zhang Y., Liu F. Isolation of Exosome Nanoparticles from Human Cerebrospinal Fluid for Proteomic Analysis. ACS Appl. Nano Mater. 2021;4:3351–3359. doi: 10.1021/acsanm.0c02622. - DOI
Grants and funding
- IP-2019-04-1511 (grant to K.G.)/Croatian Science Foundation
- uniri-biomed-18-279 (grant to M.M.), uniri-biomed-18-5 (grant to K.G.), uniri-prirod-18-299 (grant to D.K.), uniri-biomed-18-30 (grant to R.D.)/University of Rijeka
- P1-170 (grant to M.L.) and P3-054 (grant to N.K.)/Slovenian Research Agency
- Research Infrastructure for Campus-based Laboratories at the University of Rijeka project, (RC.2.2.06-0001) co-funded by the European Fund for Regional Development (ERDF) and Ministry of Science, Education and Sports of the Republic of Croatia/University of Rijeka
LinkOut - more resources
Full Text Sources
Miscellaneous

