A Platform of Patient-Derived Microtumors Identifies Individual Treatment Responses and Therapeutic Vulnerabilities in Ovarian Cancer
- PMID: 35740561
- PMCID: PMC9220902
- DOI: 10.3390/cancers14122895
A Platform of Patient-Derived Microtumors Identifies Individual Treatment Responses and Therapeutic Vulnerabilities in Ovarian Cancer
Abstract
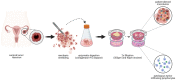
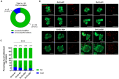
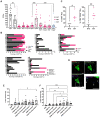
In light of the frequent development of therapeutic resistance in cancer treatment, there is a strong need for personalized model systems representing patient tumor heterogeneity, while enabling parallel drug testing and identification of appropriate treatment responses in individual patients. Using ovarian cancer as a prime example of a heterogeneous tumor disease, we developed a 3D preclinical tumor model comprised of patient-derived microtumors (PDM) and autologous tumor-infiltrating lymphocytes (TILs) to identify individual treatment vulnerabilities and validate chemo-, immuno- and targeted therapy efficacies. Enzymatic digestion of primary ovarian cancer tissue and cultivation in defined serum-free media allowed rapid and efficient recovery of PDM, while preserving histopathological features of corresponding patient tumor tissue. Reverse-phase protein array (RPPA)-analyses of >110 total and phospho-proteins enabled the identification of patient-specific sensitivities to standard, platinum-based therapy and thereby the prediction of potential treatment-responders. Co-cultures of PDM and autologous TILs for individual efficacy testing of immune checkpoint inhibitor treatment demonstrated patient-specific enhancement of cytotoxic TIL activity by this therapeutic approach. Combining protein pathway analysis and drug efficacy testing of PDM enables drug mode-of-action analyses and therapeutic sensitivity prediction within a clinically relevant time frame after surgery. Follow-up studies in larger cohorts are currently under way to further evaluate the applicability of this platform to support clinical decision making.
Keywords: RPPA protein profiling; anti-cancer drug sensitivity; cancer immunotherapy; ovarian cancer; patient-derived tumor model.
Conflict of interest statement
AH received consulting and speaking fees from GSK, AstraZeneca and Clovis. N.A., A.K., B.G., A.-L.K., A.S., S.Y.B., M.P., K.S.-L. and C.S. declare no competing interest.
Figures



Similar articles
-
Breast cancer patient-derived microtumors resemble tumor heterogeneity and enable protein-based stratification and functional validation of individualized drug treatment.J Exp Clin Cancer Res. 2023 Aug 18;42(1):210. doi: 10.1186/s13046-023-02782-2. J Exp Clin Cancer Res. 2023. PMID: 37596623 Free PMC article.
-
An autologous humanized patient-derived-xenograft platform to evaluate immunotherapy in ovarian cancer.Gynecol Oncol. 2020 Jan;156(1):222-232. doi: 10.1016/j.ygyno.2019.10.011. Epub 2019 Dec 7. Gynecol Oncol. 2020. PMID: 31818495
-
[Patient-derived microtumors : Potential for therapeutic response prediction-a case study].Urologie. 2022 Jul;61(7):739-744. doi: 10.1007/s00120-022-01851-2. Epub 2022 Jun 8. Urologie. 2022. PMID: 35925246 Review. German.
-
T cells isolated from patients with checkpoint inhibitor-resistant melanoma are functional and can mediate tumor regression.Ann Oncol. 2018 Jul 1;29(7):1575-1581. doi: 10.1093/annonc/mdy139. Ann Oncol. 2018. PMID: 29688262 Clinical Trial.
-
Tumor infiltrating lymphocytes in ovarian cancer.Cancer Biol Ther. 2015;16(6):807-20. doi: 10.1080/15384047.2015.1040960. Epub 2015 Apr 20. Cancer Biol Ther. 2015. PMID: 25894333 Free PMC article. Review.
Cited by
-
Patient-Derived Organoids as Therapy Screening Platforms in Cancer Patients.Adv Healthc Mater. 2024 Aug;13(21):e2302331. doi: 10.1002/adhm.202302331. Epub 2024 Mar 1. Adv Healthc Mater. 2024. PMID: 38359321 Free PMC article. Review.
-
Emerging roles of 3D-culture systems in tackling tumor drug resistance.Cancer Drug Resist. 2023 Nov 21;6(4):788-804. doi: 10.20517/cdr.2023.93. eCollection 2023. Cancer Drug Resist. 2023. PMID: 38263982 Free PMC article. Review.
-
Genome-wide CRISPR-Cas9 screens identify BCL family members as modulators of response to regorafenib in experimental glioma.Neuro Oncol. 2025 May 15;27(4):916-931. doi: 10.1093/neuonc/noae278. Neuro Oncol. 2025. PMID: 39756423 Free PMC article.
-
Can Nitric Oxide-Based Therapy Be Improved for the Treatment of Cancers? A Perspective.Int J Mol Sci. 2023 Sep 2;24(17):13611. doi: 10.3390/ijms241713611. Int J Mol Sci. 2023. PMID: 37686417 Free PMC article.
-
Functionally-instructed modifiers of response to ATR inhibition in experimental glioma.J Exp Clin Cancer Res. 2024 Mar 12;43(1):77. doi: 10.1186/s13046-024-02995-z. J Exp Clin Cancer Res. 2024. PMID: 38475864 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

