Epstein-Barr Virus (EBV) Is Mostly Latent and Clonal in Angioimmunoblastic T Cell Lymphoma (AITL)
- PMID: 35740565
- PMCID: PMC9221046
- DOI: 10.3390/cancers14122899
Epstein-Barr Virus (EBV) Is Mostly Latent and Clonal in Angioimmunoblastic T Cell Lymphoma (AITL)
Abstract
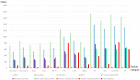
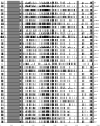
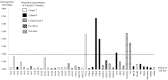
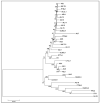
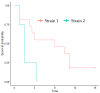
The Epstein-Barr virus (EBV) is associated with angioimmunoblastic T cell lymphoma (AITL), a peripheral T lymphoma of poor prognosis in at least 90% of cases. The role of EBV in this pathology is unknown. Using next-generation sequencing, we sequenced the entire EBV genome in biopsies from 18 patients with AITL, 16 patients with another EBV-associated lymphoma, and 2 controls. We chose an EBV target capture method, given the high specificity of this technique, followed by a second capture to increase sensitivity. We identified two main viral strains in AITL, one of them associated with the mutations BNRF1 S542N and BZLF1 A206S and with mutations in the EBNA-3 and LMP-2 genes. This strain was characterized in patients with short post-diagnosis survival. The main mutations found during AITL on the most mutated latency or tegument genes were identified and discussed. We showed that the virus was clonal in all the AITL samples, suggesting that it may be involved in this pathology. Additionally, EBV was latent in all the AITL samples; for one sample only, the virus was found to be latent and probably replicative, depending on the cells. These various elements support the role of EBV in AITL.
Keywords: AITL; EBV; Epstein–Barr virus; NGS; angioimmunoblastic T cell lymphoma; clonal; latent.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Romero-Masters J.C., Huebner S.M., Ohashi M., Bristol J.A., Benner B.E., Barlow E.A., Turk G.L., Nelson S.E., Baiu D.C., Van Sciver N., et al. B Cells infected with type 2 epstein-barr virus (EBV) have increased NFATc1/NFATc2 activity and enhanced lytic gene expression in comparison to type 1 EBV infection. PLoS Pathog. 2020;16:e1008365. doi: 10.1371/journal.ppat.1008365. - DOI - PMC - PubMed
-
- Correia S., Bridges R., Wegner F., Venturini C., Palser A., Middeldorp J.M., Cohen J.I., Lorenzetti M.A., Bassano I., White R.E., et al. Sequence variation of epstein-barr virus: Viral types, geography, codon usage, and diseases. J. Virol. 2018;92:e01132-18. doi: 10.1128/JVI.01132-18. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

