Prevalence and Prognostic Role of IDH Mutations in Acute Myeloid Leukemia: Results of the GIMEMA AML1516 Protocol
- PMID: 35740677
- PMCID: PMC9221405
- DOI: 10.3390/cancers14123012
Prevalence and Prognostic Role of IDH Mutations in Acute Myeloid Leukemia: Results of the GIMEMA AML1516 Protocol
Abstract
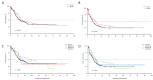
IDH1/2 mutations are common in acute myeloid leukemia (AML) and represent a therapeutic target. The GIMEMA AML1516 observational protocol was designed to study the prevalence of IDH1/2 mutations and associations with clinico-biological parameters in a cohort of Italian AML patients. We analyzed a cohort of 284 AML consecutive patients at diagnosis, 139 females and 145 males, of a median age of 65 years (range: 19−86). Of these, 38 (14%) harbored IDH1 and 51 (18%) IDH2 mutations. IDH1/2 mutations were significantly associated with WHO PS >2 (p < 0.001) and non-complex karyotype (p = 0.021) when compared to IDH1/2-WT. Furthermore, patients with IDH1 mutations were more frequently NPM1-mutated (p = 0.007) and had a higher platelet count (p = 0.036). At relapse, IDH1/2 mutations were detected in 6 (25%) patients. As per the outcome, 60.5% of IDH1/2-mutated patients achieved complete remission; overall survival and event-free survival at 2 years were 44.5% and 36.1%, respectively: these rates were similar to IDH1/2-WT. In IDH1/2-mutated patients, high WBC proved to be an independent prognostic factor for survival. In conclusion, the GIMEMA AML1516 confirms that IDH1/2 mutations are frequently detected at diagnosis and underlines the importance of recognizing IDH1/2-mutated cases up-front to offer the most appropriate therapeutic strategy, given the availability of IDH1/2 inhibitors.
Keywords: AML; DH1; IDH2; prevalence; prognosis.
Conflict of interest statement
M.V.: Amgen; Novartis, Mattioli srl; IQVIA; Dephaforum S.r.l.; M.L.: AbbVie, Acerta, Amgen, ADC Therapeutics, Astra Zeneca, BeiGene Celgene, GSKI, Gentili, Gilead/Kite, Novartis, Incyte J and J, Jazz, Regeneron, Roche, Sandoz, Takeda. Non-financial interests: PI or strategic investigator in studies supported by: Celgene, J&J, BeiGene, ADC Therapeutics; GMartinelli: Abbvie; Incyte; Pfizer; Celgene/BMS; Amgen, Roche; GlaxoSmithKline; Astellas; Daiichi Sankyo; Takeda; Janssen; Servier. Research support from: Pfizer, AbbVie, AstraZeneca, Daiichi Sankyo, Takeda, and Ariad/Incyte. GMarconi: Menarini/stemline, Pfizer, Servier, and Astellas and research support from Pfizer, AbbVie, and AstraZeneca. The other authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W. Cancer Genome Atlas Research Network, Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. - DOI - PMC - PubMed
-
- Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F., et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. - DOI - PMC - PubMed
-
- Paschka P., Schlenk R.F., Gaidzik V.I., Habdank M., Krönke J., Bullinger L., Späth D., Kayser S., Zucknick M., Götze K., et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous