Phosphatidylinositol Monophosphates Regulate the Membrane Localization of HSPA1A, a Stress-Inducible 70-kDa Heat Shock Protein
- PMID: 35740982
- PMCID: PMC9221345
- DOI: 10.3390/biom12060856
Phosphatidylinositol Monophosphates Regulate the Membrane Localization of HSPA1A, a Stress-Inducible 70-kDa Heat Shock Protein
Abstract
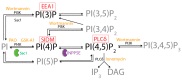
HSPA1A is a molecular chaperone that regulates the survival of stressed and cancer cells. In addition to its cytosolic pro-survival functions, HSPA1A also localizes and embeds in the plasma membrane (PM) of stressed and tumor cells. Membrane-associated HSPA1A exerts immunomodulatory functions and renders tumors resistant to standard therapies. Therefore, understanding and manipulating HSPA1A's surface presentation is a promising therapeutic. However, HSPA1A's pathway to the cell surface remains enigmatic because this protein lacks known membrane localization signals. Considering that HSPA1A binds to lipids, like phosphatidylserine (PS) and monophosphorylated phosphoinositides (PIPs), we hypothesized that this interaction regulates HSPA1A's PM localization and anchorage. To test this hypothesis, we subjected human cell lines to heat shock, depleted specific lipid targets, and quantified HSPA1A's PM localization using confocal microscopy and cell surface biotinylation. These experiments revealed that co-transfection of HSPA1A with lipid-biosensors masking PI(4)P and PI(3)P significantly reduced HSPA1A's heat-induced surface presentation. Next, we manipulated the cellular lipid content using ionomycin, phenyl arsine oxide (PAO), GSK-A1, and wortmannin. These experiments revealed that HSPA1A's PM localization was unaffected by ionomycin but was significantly reduced by PAO, GSK-A1, and wortmannin, corroborating the findings obtained by the co-transfection experiments. We verified these results by selectively depleting PI(4)P and PI(4,5)P2 using a rapamycin-induced phosphatase system. Our findings strongly support the notion that HSPA1A's surface presentation is a multifaceted lipid-driven phenomenon controlled by the binding of the chaperone to specific endosomal and PM lipids.
Keywords: cell surface; heat-shock proteins; lipids; pharmacology; phosphoinositide; plasma membrane.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Membrane Localization of HspA1A, a Stress Inducible 70-kDa Heat-Shock Protein, Depends on Its Interaction with Intracellular Phosphatidylserine.Biomolecules. 2019 Apr 17;9(4):152. doi: 10.3390/biom9040152. Biomolecules. 2019. PMID: 30999671 Free PMC article.
-
Characterization of the Relationship between the Chaperone and Lipid-Binding Functions of the 70-kDa Heat-Shock Protein, HspA1A.Int J Mol Sci. 2020 Aug 20;21(17):5995. doi: 10.3390/ijms21175995. Int J Mol Sci. 2020. PMID: 32825419 Free PMC article.
-
Heat-Induced Phosphatidylserine Changes Drive HSPA1A's Plasma Membrane Localization.bioRxiv [Preprint]. 2024 Dec 11:2024.12.02.626454. doi: 10.1101/2024.12.02.626454. bioRxiv. 2024. Update in: Cell Stress Chaperones. 2025 Jul 11;30(5):100092. doi: 10.1016/j.cstres.2025.100092. PMID: 39713339 Free PMC article. Updated. Preprint.
-
Distinguishing integral and receptor-bound heat shock protein 70 (Hsp70) on the cell surface by Hsp70-specific antibodies.Cell Stress Chaperones. 2011 May;16(3):251-5. doi: 10.1007/s12192-010-0247-1. Epub 2010 Dec 17. Cell Stress Chaperones. 2011. PMID: 21165727 Free PMC article. Review.
-
Hsp70 interactions with membrane lipids regulate cellular functions in health and disease.Prog Lipid Res. 2019 Apr;74:18-30. doi: 10.1016/j.plipres.2019.01.004. Epub 2019 Jan 30. Prog Lipid Res. 2019. PMID: 30710597 Review.
Cited by
-
Heat Shock-Induced PI(4)P Increase Drives HSPA1A Translocation to the Plasma Membrane in Cancer and Stressed Cells through PI4KIII Alpha Activation.bioRxiv [Preprint]. 2025 Feb 20:2025.02.16.638537. doi: 10.1101/2025.02.16.638537. bioRxiv. 2025. PMID: 40027828 Free PMC article. Preprint.
-
Chronotherapeutic neuroprotective effect of verapamil against lipopolysaccharide-induced neuroinflammation in mice through modulation of calcium-dependent genes.Mol Med. 2022 Nov 26;28(1):139. doi: 10.1186/s10020-022-00564-8. Mol Med. 2022. PMID: 36435759 Free PMC article.
-
Exploring the Role of T-Cell Metabolism in Modulating Immunotherapy Efficacy for Non-Small Cell Lung Cancer Based on Clustering.J Clin Lab Anal. 2025 Jul;39(13):e25020. doi: 10.1002/jcla.25020. Epub 2025 Apr 17. J Clin Lab Anal. 2025. PMID: 40244859 Free PMC article.
-
PDT-regulated immune gene prognostic model reveals tumor microenvironment in colorectal cancer liver metastases.Sci Rep. 2025 Apr 16;15(1):13129. doi: 10.1038/s41598-025-97667-z. Sci Rep. 2025. PMID: 40240471 Free PMC article.
-
Closest horizons of Hsp70 engagement to manage neurodegeneration.Front Mol Neurosci. 2023 Sep 19;16:1230436. doi: 10.3389/fnmol.2023.1230436. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37795273 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

