BECLIN-1-Mediated Autophagy Suppresses Silica Nanoparticle-Induced Testicular Toxicity via the Inhibition of Caspase 8-Mediated Cell Apoptosis in Leydig Cells
- PMID: 35740992
- PMCID: PMC9221084
- DOI: 10.3390/cells11121863
BECLIN-1-Mediated Autophagy Suppresses Silica Nanoparticle-Induced Testicular Toxicity via the Inhibition of Caspase 8-Mediated Cell Apoptosis in Leydig Cells
Abstract
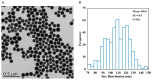
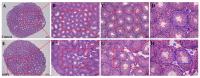
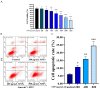
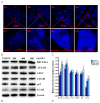
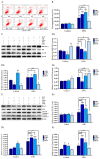
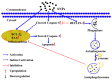
Accumulation of silica nanoparticles (SNPs) in the testes leads to male reproductive toxicity. However, little is known about the effect and mechanistic insights of SNP-induced autophagy on apoptosis in Leydig cells. In this study, we aimed to verify the role of SNP-induced autophagy in apoptosis and explore the possible underlying mechanism in mouse primary Leydig cells (PLCs). H&E staining showed that SNPs changed the histological structures of the testes, including a reduction in the Leydig cell populations in vivo. CCK-8 assay showed that SNPs decreased cell viability, and flow cytometry showed that SNPs increased cell apoptosis, both in a dose-dependent manner in vitro. Additionally, Western blotting further found that SNPs activated autophagy by an increase in BECLIN-1, ATG16L, and LC3-II levels and promoted the intrinsic pathway of apoptosis by an increase in the BAX/BCL-2 ratio, cleaved the caspase 8 and caspase 3 levels. Furthermore, autophagy decreased SNP-induced apoptosis via regulation of the caspase 8 level combined with rapamycin, 3-methyladenine, and chloroquine. BECLIN-1 depletion increased the caspase 8 level, leading to an increase in SNP-induced cell apoptosis. Collectively, this evidence demonstrates that SNPs activated BECLIN-1-mediated autophagy, which prevented SNP-induced testicular toxicity via the inhibition of caspase 8-mediated cell apoptosis in Leydig cells.
Keywords: BECLIN-1; Leydig cells; apoptosis; autophagy; silica nanoparticles; testicular toxicity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

