Lung Spatial Profiling Reveals a T Cell Signature in COPD Patients with Fatal SARS-CoV-2 Infection
- PMID: 35740993
- PMCID: PMC9220844
- DOI: 10.3390/cells11121864
Lung Spatial Profiling Reveals a T Cell Signature in COPD Patients with Fatal SARS-CoV-2 Infection
Abstract
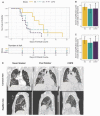
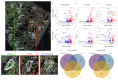
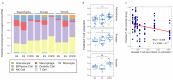
People with pre-existing lung diseases such as chronic obstructive pulmonary disease (COPD) are more likely to get very sick from SARS-CoV-2 disease 2019 (COVID-19). Still, an interrogation of the immune response to COVID-19 infection, spatially throughout the lung structure, is lacking in patients with COPD. For this study, we characterized the immune microenvironment of the lung parenchyma, airways, and vessels of never- and ever-smokers with or without COPD, all of whom died of COVID-19, using spatial transcriptomic and proteomic profiling. The parenchyma, airways, and vessels of COPD patients, compared to control lungs had (1) significant enrichment for lung-resident CD45RO+ memory CD4+ T cells; (2) downregulation of genes associated with T cell antigen priming and memory T cell differentiation; and (3) higher expression of proteins associated with SARS-CoV-2 entry and primary receptor ubiquitously across the ROIs and in particular the lung parenchyma, despite similar SARS-CoV-2 structural gene expression levels. In conclusion, the lung parenchyma, airways, and vessels of COPD patients have increased T-lymphocytes with a blunted memory CD4 T cell response and a more invasive SARS-CoV-2 infection pattern and may underlie the higher death toll observed with COVID-19.
Keywords: COVID-19; T cells; chronic obstructive pulmonary disease (COPD).
Conflict of interest statement
No conflicts of interest to declare.
Figures


References
-
- COVID-19. Corona Virus Pandemic. [(accessed on 1 January 2022)]. Available online: https://www.worldometers.info/coronavirus/
-
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

