Diagnostics of COVID-19 Based on CRISPR-Cas Coupled to Isothermal Amplification: A Comparative Analysis and Update
- PMID: 35741243
- PMCID: PMC9222122
- DOI: 10.3390/diagnostics12061434
Diagnostics of COVID-19 Based on CRISPR-Cas Coupled to Isothermal Amplification: A Comparative Analysis and Update
Abstract
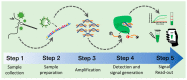
The emergence of the COVID-19 pandemic prompted fast development of novel diagnostic methods of the etiologic virus SARS-CoV-2. Methods based on CRISPR-Cas systems have been particularly promising because they can achieve a similar sensitivity and specificity to the benchmark RT-qPCR, especially when coupled to an isothermal pre-amplification step. Furthermore, they have also solved inherent limitations of RT-qPCR that impede its decentralized use and deployment in the field, such as the need for expensive equipment, high cost per reaction, and delivery of results in hours, among others. In this review, we evaluate publicly available methods to detect SARS-CoV-2 that are based on CRISPR-Cas and isothermal amplification. We critically analyze the steps required to obtain a successful result from clinical samples and pinpoint key experimental conditions and parameters that could be optimized or modified to improve clinical and analytical outputs. The COVID outbreak has propelled intensive research in a short time, which is paving the way to develop effective and very promising CRISPR-Cas systems for the precise detection of SARS-CoV-2. This review could also serve as an introductory guide to new labs delving into this technology.
Keywords: CRISPR–Cas; SARS-CoV-2; comparative analysis; isothermal amplification; molecular diagnostics; nucleic acid detection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Johns Hopkins Coronavirus Resource Center COVID-19 Map. [(accessed on 29 March 2022)]. Available online: https://coronavirus.jhu.edu/map.html.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

