Different Lung Parenchyma Quantification Using Dissimilar Segmentation Software: A Multi-Center Study for COVID-19 Patients
- PMID: 35741310
- PMCID: PMC9222070
- DOI: 10.3390/diagnostics12061501
Different Lung Parenchyma Quantification Using Dissimilar Segmentation Software: A Multi-Center Study for COVID-19 Patients
Abstract
Background: Chest Computed Tomography (CT) imaging has played a central role in the diagnosis of interstitial pneumonia in patients affected by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and can be used to obtain the extent of lung involvement in COVID-19 pneumonia patients either qualitatively, via visual inspection, or quantitatively, via AI-based software. This study aims to compare the qualitative/quantitative pathological lung extension data on COVID-19 patients. Secondly, the quantitative data obtained were compared to verify their concordance since they were derived from three different lung segmentation software.
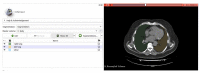
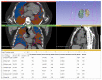
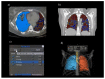
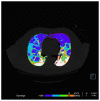
Methods: This double-center study includes a total of 120 COVID-19 patients (60 from each center) with positive reverse-transcription polymerase chain reaction (RT-PCR) who underwent a chest CT scan from November 2020 to February 2021. CT scans were analyzed retrospectively and independently in each center. Specifically, CT images were examined manually by two different and experienced radiologists for each center, providing the qualitative extent score of lung involvement, whereas the quantitative analysis was performed by one trained radiographer for each center using three different software: 3DSlicer, CT Lung Density Analysis, and CT Pulmo 3D.
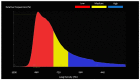
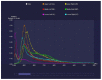
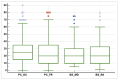
Results: The agreement between radiologists for visual estimation of pneumonia at CT can be defined as good (ICC 0.79, 95% CI 0.73-0.84). The statistical tests show that 3DSlicer overestimates the measures assessed; however, ICC index returns a value of 0.92 (CI 0.90-0.94), indicating excellent reliability within the three software employed. ICC was also performed between each single software and the median of the visual score provided by the radiologists. This statistical analysis underlines that the best agreement is between 3D Slicer "LungCTAnalyzer" and the median of the visual score (0.75 with a CI 0.67-82 and with a median value of 22% of disease extension for the software and 25% for the visual values).
Conclusions: This study provides for the first time a direct comparison between the actual gold standard, which is represented by the qualitative information described by radiologists, and novel quantitative AI-based techniques, here represented by three different commonly used lung segmentation software, underlying the importance of these specific values that in the future could be implemented as consistent prognostic and clinical course parameters.
Keywords: COVID-19 pneumonia; chest CT; lung segmentation; post-processing tools; semi-automatic segmentation software.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Al World Health Organization COVID-19—China. 2020. [(accessed on 5 December 2021)]. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2020-DON233.
-
- World Health Organization WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. 2020. [(accessed on 5 December 2021)]. Available online: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-re....
LinkOut - more resources
Full Text Sources
Miscellaneous

