NeDSeM: Neutrosophy Domain-Based Segmentation Method for Malignant Melanoma Images
- PMID: 35741504
- PMCID: PMC9222744
- DOI: 10.3390/e24060783
NeDSeM: Neutrosophy Domain-Based Segmentation Method for Malignant Melanoma Images
Abstract
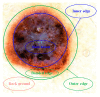
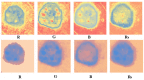
Skin lesion segmentation is the first and indispensable step of malignant melanoma recognition and diagnosis. At present, most of the existing skin lesions segmentation techniques often used traditional methods like optimum thresholding, etc., and deep learning methods like U-net, etc. However, the edges of skin lesions in malignant melanoma images are gradually changed in color, and this change is nonlinear. The existing methods can not effectively distinguish banded edges between lesion areas and healthy skin areas well. Aiming at the uncertainty and fuzziness of banded edges, the neutrosophic set theory is used in this paper which is better than fuzzy theory to deal with banded edge segmentation. Therefore, we proposed a neutrosophy domain-based segmentation method that contains six steps. Firstly, an image is converted into three channels and the pixel matrix of each channel is obtained. Secondly, the pixel matrixes are converted into Neutrosophic Set domain by using the neutrosophic set conversion method to express the uncertainty and fuzziness of banded edges of malignant melanoma images. Thirdly, a new Neutrosophic Entropy model is proposed to combine the three memberships according to some rules by using the transformations in the neutrosophic space to comprehensively express three memberships and highlight the banded edges of the images. Fourthly, the feature augment method is established by the difference of three components. Fifthly, the dilation is used on the neutrosophic entropy matrixes to fill in the noise region. Finally, the image that is represented by transformed matrix is segmented by the Hierarchical Gaussian Mixture Model clustering method to obtain the banded edge of the image. Qualitative and quantitative experiments are performed on malignant melanoma image dataset to evaluate the performance of the NeDSeM method. Compared with some state-of-the-art methods, our method has achieved good results in terms of performance and accuracy.
Keywords: HGMM; image segmentation; malignant melanoma image; morphology; neutrosophic entropy; neutrosophic set.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures









Similar articles
-
Automatic segmentation of tumors in B-Mode breast ultrasound images using information gain based neutrosophic clustering.J Xray Sci Technol. 2018;26(2):209-225. doi: 10.3233/XST-17313. J Xray Sci Technol. 2018. PMID: 29154313
-
A neutrosophic-entropy based clustering algorithm (NEBCA) with HSV color system: A special application in segmentation of Parkinson's disease (PD) MR images.Comput Methods Programs Biomed. 2020 Jun;189:105317. doi: 10.1016/j.cmpb.2020.105317. Epub 2020 Jan 14. Comput Methods Programs Biomed. 2020. PMID: 31981758
-
A neutrosophic-entropy based adaptive thresholding segmentation algorithm: A special application in MR images of Parkinson's disease.Artif Intell Med. 2020 Apr;104:101838. doi: 10.1016/j.artmed.2020.101838. Epub 2020 Feb 28. Artif Intell Med. 2020. PMID: 32499006
-
Deep Learning Approaches Towards Skin Lesion Segmentation and Classification from Dermoscopic Images - A Review.Curr Med Imaging. 2020;16(5):513-533. doi: 10.2174/1573405615666190129120449. Curr Med Imaging. 2020. PMID: 32484086 Review.
-
Deep learning-based automatic segmentation of images in cardiac radiography: A promising challenge.Comput Methods Programs Biomed. 2022 Jun;220:106821. doi: 10.1016/j.cmpb.2022.106821. Epub 2022 Apr 19. Comput Methods Programs Biomed. 2022. PMID: 35487181 Review.
Cited by
-
Bendlet Transform Based Adaptive Denoising Method for Microsection Images.Entropy (Basel). 2022 Jun 24;24(7):869. doi: 10.3390/e24070869. Entropy (Basel). 2022. PMID: 35885092 Free PMC article.
References
-
- Celebi M.E., Wen Q.U., Iyatomi H.I., Shimizu K.O., Zhou H., Schaefer G. A state-of-the-art survey on lesion border detection in dermoscopy images. Dermoscopy Image Anal. 2015;10:97–129.
Grants and funding
LinkOut - more resources
Full Text Sources

