Clinical Targeted Panel Sequencing Analysis in Clinical Evaluation of Children with Autism Spectrum Disorder in China
- PMID: 35741772
- PMCID: PMC9222325
- DOI: 10.3390/genes13061010
Clinical Targeted Panel Sequencing Analysis in Clinical Evaluation of Children with Autism Spectrum Disorder in China
Abstract
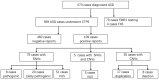
Autism spectrum disorder (ASD) is an early-onset neurodevelopmental disorder in which genetics play a major role. Molecular diagnosis may lead to a more accurate prognosis, improved clinical management, and potential treatment of the condition. Both copy number variations (CNVs) and single nucleotide variations (SNVs) have been reported to contribute to the genetic etiology of ASD. The effectiveness and validity of clinical targeted panel sequencing (CTPS) designed to analyze both CNVs and SNVs can be evaluated in different ASD cohorts. CTPS was performed on 573 patients with the diagnosis of ASD. Medical records of positive CTPS cases were further reviewed and analyzed. Additional medical examinations were performed for a group of selective cases. Positive molecular findings were confirmed by orthogonal methods. The overall positive rate was 19.16% (109/569) in our cohort. About 13.89% (79/569) and 4.40% (25/569) of cases had SNVs only and CNVs only findings, respectively, while 0.9% (5/569) of cases had both SNV and CNV findings. For cases with SNVs findings, the SHANK3 gene has the greatest number of reportable variants, followed by gene MYT1L. Patients with MYT1L variants share common and specific clinical characteristics. We found a child with compound heterozygous SLC26A4 variants had an enlarged vestibular aqueduct syndrome and autistic phenotype. Our results showed that CTPS is an effective molecular diagnostic tool for ASD. Thorough clinical and genetic evaluation of ASD can lead to more accurate diagnosis and better management of the condition.
Keywords: MYT1L; SLC26A4; autism spectrum disorder; genetic variants; targeted panel sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Targeted sequencing and clinical strategies in children with autism spectrum disorder: A cohort study.Front Genet. 2023 Feb 28;14:1083779. doi: 10.3389/fgene.2023.1083779. eCollection 2023. Front Genet. 2023. PMID: 37007974 Free PMC article.
-
Impact of on-site clinical genetics consultations on diagnostic rate in children and young adults with autism spectrum disorder.Mol Autism. 2019 Aug 7;10:33. doi: 10.1186/s13229-019-0284-2. eCollection 2019. Mol Autism. 2019. PMID: 31406558 Free PMC article.
-
Genome-wide analysis of copy number variations identifies PARK2 as a candidate gene for autism spectrum disorder.Mol Autism. 2016 Apr 1;7:23. doi: 10.1186/s13229-016-0087-7. eCollection 2016. Mol Autism. 2016. PMID: 27042285 Free PMC article.
-
Syndromic autism spectrum disorders: moving from a clinically defined to a molecularly defined approach.Dialogues Clin Neurosci. 2017 Dec;19(4):353-371. doi: 10.31887/DCNS.2017.19.4/sscherer. Dialogues Clin Neurosci. 2017. PMID: 29398931 Free PMC article. Review.
-
Autism Spectrum Disorder: Genetic Mechanisms and Inheritance Patterns.Genes (Basel). 2025 Apr 23;16(5):478. doi: 10.3390/genes16050478. Genes (Basel). 2025. PMID: 40428300 Free PMC article. Review.
Cited by
-
Molecular genetic testing and cohort analysis of 32 twin pairs with neurodevelopmental disorders-Reporting a novel de novo variant of TET3.Hum Genomics. 2025 Apr 21;19(1):42. doi: 10.1186/s40246-025-00748-3. Hum Genomics. 2025. PMID: 40259394 Free PMC article.
-
Biogenic Amine Metabolism and Its Genetic Variations in Autism Spectrum Disorder: A Comprehensive Overview.Biomolecules. 2025 Apr 7;15(4):539. doi: 10.3390/biom15040539. Biomolecules. 2025. PMID: 40305279 Free PMC article. Review.
-
Identification of copy number variants with genome sequencing: Clinical experiences from the NYCKidSeq program.Clin Genet. 2023 Aug;104(2):210-225. doi: 10.1111/cge.14365. Epub 2023 Jun 19. Clin Genet. 2023. PMID: 37334874 Free PMC article.
-
NGS Custom Panel Implementation in Patients with Non-Syndromic Autism Spectrum Disorders in the Clinical Routine of a Tertiary Hospital.Genes (Basel). 2023 Nov 17;14(11):2091. doi: 10.3390/genes14112091. Genes (Basel). 2023. PMID: 38003033 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

