Tissue-Specific and Time-Dependent Expressions of PC4s in Bay Scallop (Argopecten irradians irradians) Reveal Function Allocation in Thermal Response
- PMID: 35741819
- PMCID: PMC9223095
- DOI: 10.3390/genes13061057
Tissue-Specific and Time-Dependent Expressions of PC4s in Bay Scallop (Argopecten irradians irradians) Reveal Function Allocation in Thermal Response
Abstract
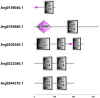
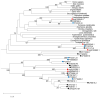
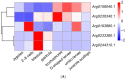
Transcriptional coactivator p15 (PC4) encodes a structurally conserved but functionally diverse protein that plays crucial roles in RNAP-II-mediated transcription, DNA replication and damage repair. Although structures and functions of PC4 have been reported in most vertebrates and some invertebrates, the PC4 genes were less systematically identified and characterized in the bay scallop Argopecten irradians irradians. In this study, five PC4 genes (AiPC4s) were successfully identified in bay scallops via whole-genome scanning through in silico analysis. Protein structure and phylogenetic analyses of AiPC4s were conducted to determine the identities and evolutionary relationships of these genes. Expression levels of AiPC4s were assessed in embryos/larvae at all developmental stages, in healthy adult tissues and in different tissues (mantles, gills, hemocytes and hearts) being processed under 32 °C stress with different time durations (0 h, 6 h, 12 h, 24 h, 3 d, 6 d and 10 d). Spatiotemporal expression profiles of AiPC4s suggested the functional roles of the genes in embryos/larvae at all developmental stages and in healthy adult tissues in bay scallop. Expression regulations (up- and down-) of AiPC4s under high-temperature stress displayed both tissue-specific and time-dependent patterns with function allocations, revealing that AiPC4s performed differentiated functions in response to thermal stress. This work provides clues of molecular function allocation of PC4 in scallops in response to thermal stress and helps in illustrating how marine bivalves resist elevated seawater temperature.
Keywords: Argopecten irradians irradians; expression regulation; function allocation; thermal tolerance; transcriptional coactivator p15.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Masson-Delmotte V., Zhai P., Chen Y., Goldfarb L., Gomis M.I., Matthews J.B.R., Berger S., Huang M., Yelekçi O., Yu R., et al. Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change. IPCC; Geneva, Switzerland: 2021. Summary for Policymakers.
-
- Graham N.A.J., McClanahan T.R., MacNeil M.A., Wilson S.K., Polunin N.V.C., Jennings S., Chabanet P., Clark S., Spalding M.D., Letourneur Y., et al. Climate warming, marine protected areas and the ocean-scale integrity of coral reef ecosystems. PLoS ONE. 2008;3:e3039. doi: 10.1371/journal.pone.0003039. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

