Characterization of Immune-Based Molecular Subtypes and Prognostic Model in Prostate Adenocarcinoma
- PMID: 35741849
- PMCID: PMC9223199
- DOI: 10.3390/genes13061087
Characterization of Immune-Based Molecular Subtypes and Prognostic Model in Prostate Adenocarcinoma
Abstract
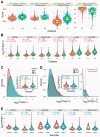
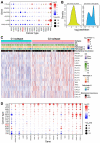
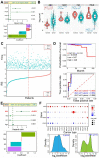
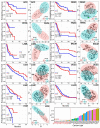
Prostate adenocarcinoma (PRAD), also named prostate cancer, the most common visceral malignancy, is diagnosed in male individuals. Herein, in order to obtain immune-based subtypes, we performed an integrative analysis to characterize molecular subtypes based on immune-related genes, and further discuss the potential features and differences between identified subtypes. Simultaneously, we also construct an immune-based risk model to assess cancer prognosis. Our findings showed that the two subtypes, C1 and C2, could be characterized, and the two subtypes showed different characteristics that could clearly describe the heterogeneity of immune microenvironments. The C2 subtype presented a better survival rate than that in the C1 subtype. Further, we constructed an immune-based prognostic model based on four screened abnormally expressed genes, and they were selected as predictors of the robust prognostic model (AUC = 0.968). Our studies provide reference for characterization of molecular subtypes and immunotherapeutic agents against prostate cancer, and the developed robust and useful immune-based prognostic model can contribute to cancer prognosis and provide reference for the individualized treatment plan and health resource utilization. These findings further promote the development and application of precision medicine in prostate cancer.
Keywords: immune-based; molecular subtypes; prognostic model; prostate adenocarcinoma (PRAD).
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

