The Identification of Broomcorn Millet bZIP Transcription Factors, Which Regulate Growth and Development to Enhance Stress Tolerance and Seed Germination
- PMID: 35742892
- PMCID: PMC9224411
- DOI: 10.3390/ijms23126448
The Identification of Broomcorn Millet bZIP Transcription Factors, Which Regulate Growth and Development to Enhance Stress Tolerance and Seed Germination
Abstract
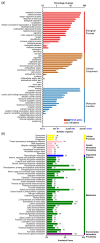
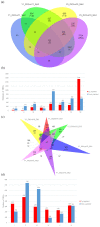
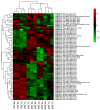
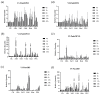
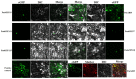
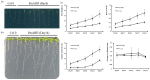
Broomcorn millet (Panicum miliaceum L.) is a water-efficient and highly salt-tolerant plant. In this study, the salt tolerance of 17 local species of broomcorn millet was evaluated through testing based on the analysis of the whitening time and the germination rate of their seeds. Transcriptome sequencing revealed that PmbZIP131, PmbZIP125, PmbZIP33, PmABI5, PmbZIP118, and PmbZIP97 are involved in seed germination under salt stress. Seedling stage expression analysis indicates that PmABI5 expression was induced by treatments of high salt (200 mM NaCl), drought (20% W/V PEG6000), and low temperature (4 °C) in seedlings of the salt-tolerant variety Y9. The overexpression of PmABI5 significantly increases the germination rate and root traits of Arabidopsis thaliana transgenic lines, with root growth and grain traits significantly enhanced compared to the wild type (Nipponbare). BiFC showed that PmABI5 undergoes homologous dimerization in addition to forming a heterodimer with either PmbZIP33 or PmbZIP131. Further yeast one-hybrid experiments showed that PmABI5 and PmbZIP131 regulate the expression of PmNAC1 by binding to the G-box in the promoter. These results indicate that PmABI5 can directly regulate seed germination and seedling growth and indirectly improve the salt tolerance of plants by regulating the expression of the PmNAC1 gene through the formation of heterodimers with PmbZIP131.
Keywords: NAC transcription factor; bZIP transcription factor; gene family analysis; root development; stress response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- FAOSTAT Food Balance. 1993–2013 and 2010–2019. [(accessed on 16 November 2021)]. Available online: https://www.fao.org/faostat/en/#data/FBSH.
-
- Ritchie H., Roser M. Crop Yields. Our World in Data. 2018. [(accessed on 16 November 2021)]. Available online: https://ourworldindata.org/
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

