A Study of Drug Repurposing to Identify SARS-CoV-2 Main Protease (3CLpro) Inhibitors
- PMID: 35742913
- PMCID: PMC9224295
- DOI: 10.3390/ijms23126468
A Study of Drug Repurposing to Identify SARS-CoV-2 Main Protease (3CLpro) Inhibitors
Abstract
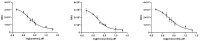
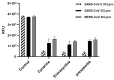
The outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) wreaked havoc all over the world. Although vaccines for the disease have recently become available and started to be administered to the population in various countries, there is still a strong and urgent need for treatments to cure COVID-19. One of the safest and fastest strategies is represented by drug repurposing (DRPx). In this study, thirty compounds with known safety profiles were identified from a chemical library of Phase II-and-up compounds through a combination of SOM Biotech's Artificial Intelligence (AI) technology, SOMAIPRO, and in silico docking calculations with third-party software. The selected compounds were then tested in vitro for inhibitory activity against SARS-CoV-2 main protease (3CLpro or Mpro). Of the thirty compounds, three (cynarine, eravacycline, and prexasertib) displayed strong inhibitory activity against SARS-CoV-2 3CLpro. VeroE6 cells infected with SARS-CoV-2 were used to find the cell protection capability of each candidate. Among the three compounds, only eravacycline showed potential antiviral activities with no significant cytotoxicity. A further study is planned for pre-clinical trials.
Keywords: SARS-CoV-2 3CL protease; antiviral; drug repurposing; fret; inhibitory compounds.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

