Integrative Proteome and Phosphoproteome Profiling of Early Cold Response in Maize Seedlings
- PMID: 35742945
- PMCID: PMC9224472
- DOI: 10.3390/ijms23126493
Integrative Proteome and Phosphoproteome Profiling of Early Cold Response in Maize Seedlings
Abstract
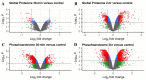
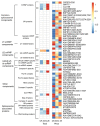
Cold limits the growth and yield of maize in temperate regions, but the molecular mechanism of cold adaptation remains largely unexplored in maize. To identify early molecular events during cold shock, maize seedlings were treated under 4 °C for 30 min and 2 h, and analyzed at both the proteome and phosphoproteome levels. Over 8500 proteins and 19,300 phosphopeptides were quantified. About 660 and 620 proteins were cold responsive at protein abundance or site-specific phosphorylation levels, but only 65 proteins were shared between them. Functional enrichment analysis of cold-responsive proteins and phosphoproteins revealed that early cold response in maize is associated with photosynthesis light reaction, spliceosome, endocytosis, and defense response, consistent with similar studies in Arabidopsis. Thirty-two photosynthesis proteins were down-regulated at protein levels, and 48 spliceosome proteins were altered at site-specific phosphorylation levels. Thirty-one kinases and 33 transcriptional factors were cold responsive at protein, phosphopeptide, or site-specific phosphorylation levels. Our results showed that maize seedlings respond to cold shock rapidly, at both the proteome and phosphoproteome levels. This study provides a comprehensive landscape at the cold-responsive proteome and phosphoproteome in maize seedlings that can be a significant resource to understand how C4 plants respond to a sudden temperature drop.
Keywords: TMT-labeling; cold stress; maize; phosphoproteome; photosynthesis; proteome; seedlings; spliceosome.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures







References
-
- Frascaroli E., Revilla P. The Maize Genome. Springer; Cham, Switzerland: 2018. Genomics of cold tolerance in maize; pp. 287–303.
-
- Farooq M., Aziz T., Wahid A., Lee D.J., Siddique K.H.M. Chilling tolerance in maize: Agronomic and physiological approaches. Crop. Pasture Sci. 2009;60:501–516. doi: 10.1071/CP08427. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

