GWAS Reveals a Novel Candidate Gene CmoAP2/ERF in Pumpkin (Cucurbita moschata) Involved in Resistance to Powdery Mildew
- PMID: 35742978
- PMCID: PMC9223685
- DOI: 10.3390/ijms23126524
GWAS Reveals a Novel Candidate Gene CmoAP2/ERF in Pumpkin (Cucurbita moschata) Involved in Resistance to Powdery Mildew
Abstract
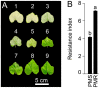
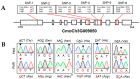
Pumpkin (Cucurbita moschata Duchesne ex Poir.) is a multipurpose cash crop rich in antioxidants, minerals, and vitamins; the seeds are also a good source of quality oils. However, pumpkin is susceptible to the fungus Podosphaera xanthii, an obligate biotrophic pathogen, which usually causes powdery mildew (PM) on both sides of the leaves and reduces photosynthesis. The fruits of infected plants are often smaller than usual and unpalatable. This study identified a novel gene that involves PM resistance in pumpkins through a genome-wide association study (GWAS). The allelic variation identified in the CmoCh3G009850 gene encoding for AP2-like ethylene-responsive transcription factor (CmoAP2/ERF) was proven to be involved in PM resistance. Validation of the GWAS data revealed six single nucleotide polymorphism (SNP) variations in the CmoAP2/ERF coding sequence between the resistant (IT 274039 [PMR]) and the susceptible (IT 278592 [PMS]). A polymorphic marker (dCAPS) was developed based on the allelic diversity to differentiate these two haplotypes. Genetic analysis in the segregating population derived from PMS and PMR parents provided evidence for an incomplete dominant gene-mediated PM resistance. Further, the qRT-PCR assay validated the elevated expression of CmoAP2/ERF during PM infection in the PMR compared with PMS. These results highlighted the pivotal role of CmoAP2/ERF in conferring resistance to PM and identifies it as a valuable molecular entity for breeding resistant pumpkin cultivars.
Keywords: CmoAP2/ERF; Cucurbita moschata Duchesne ex Poir.; Podosphaera xanthii; genome-wide association study; powdery mildew; powdery mildew resistance; pumpkin; single nucleotide polymorphism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Lee S.-C., Lee W.-K., Ali A., Kumar M., Yang T.-J., Song K. Genome-wide identification and classification of the AP2/EREBP gene family in the Cucurbitaceae species. Plant Breed. Biotechnol. 2017;5:122–133. doi: 10.9787/PBB.2017.5.2.123. - DOI
-
- Bisognin D.A. Origin and evolution of cultivated cucurbits. Cienc. Rural. 2002;32:715–723. doi: 10.1590/S0103-84782002000400028. - DOI
-
- Dhiman A.K., Sharma K.D., Attri S. Functional constituents and processing of pumpkin: A review. J. Food Sci. Technol. 2009;46:411–417.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

