Distinct Roles of NANOS1 and NANOS3 in the Cell Cycle and NANOS3-PUM1-FOXM1 Axis to Control G2/M Phase in a Human Primordial Germ Cell Model
- PMID: 35743036
- PMCID: PMC9223905
- DOI: 10.3390/ijms23126592
Distinct Roles of NANOS1 and NANOS3 in the Cell Cycle and NANOS3-PUM1-FOXM1 Axis to Control G2/M Phase in a Human Primordial Germ Cell Model
Abstract
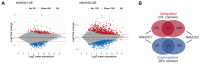
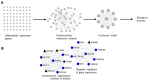
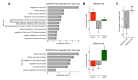
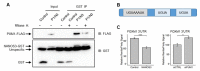
Nanos RNA-binding proteins are critical factors of germline development throughout the animal kingdom and their dysfunction causes infertility. During evolution, mammalian Nanos paralogues adopted divergent roles in germ cell biology. However, the molecular basis behind this divergence, such as their target mRNAs, remains poorly understood. Our RNA-sequencing analysis in a human primordial germ cell model-TCam-2 cell line revealed distinct pools of genes involved in the cell cycle process downregulated upon NANOS1 and NANOS3 overexpression. We show that NANOS1 and NANOS3 proteins influence different stages of the cell cycle. Namely, NANOS1 is involved in the G1/S and NANOS3 in the G2/M phase transition. Many of their cell cycle targets are known infertility and cancer-germ cell genes. Moreover, NANOS3 in complex with RNA-binding protein PUM1 causes 3'UTR-mediated repression of FOXM1 mRNA encoding a transcription factor crucial for G2/M phase transition. Interestingly, while NANOS3 and PUM1 act as post-transcriptional repressors of FOXM1, FOXM1 potentially acts as a transcriptional activator of NANOS3, PUM1, and itself. Finally, by utilizing publicly available RNA-sequencing datasets, we show that the balance between FOXM1-NANOS3 and FOXM1-PUM1 expression levels is disrupted in testis cancer, suggesting a potential role in this disease.
Keywords: FOXM1; NANOS1; NANOS3; PUM1; cell cycle; germ cell cancer; human primordial germ cells.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures




Similar articles
-
Human NANOS1 Represses Apoptosis by Downregulating Pro-Apoptotic Genes in the Male Germ Cell Line.Int J Mol Sci. 2020 Apr 24;21(8):3009. doi: 10.3390/ijms21083009. Int J Mol Sci. 2020. PMID: 32344590 Free PMC article.
-
PUM1 and PUM2 exhibit different modes of regulation for SIAH1 that involve cooperativity with NANOS paralogues.Cell Mol Life Sci. 2019 Jan;76(1):147-161. doi: 10.1007/s00018-018-2926-5. Epub 2018 Sep 29. Cell Mol Life Sci. 2019. PMID: 30269240 Free PMC article.
-
Primordial germ cell identification and traceability during the initial development of the Siluriformes fish Pseudopimelodus mangurus.Fish Physiol Biochem. 2022 Oct;48(5):1137-1153. doi: 10.1007/s10695-022-01106-z. Epub 2022 Aug 4. Fish Physiol Biochem. 2022. PMID: 35925505
-
Nanos genes and their role in development and beyond.Cell Mol Life Sci. 2018 Jun;75(11):1929-1946. doi: 10.1007/s00018-018-2766-3. Epub 2018 Feb 3. Cell Mol Life Sci. 2018. PMID: 29397397 Free PMC article. Review.
-
The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles.Adv Cancer Res. 2013;118:97-398. doi: 10.1016/B978-0-12-407173-5.00004-2. Adv Cancer Res. 2013. PMID: 23768511 Review.
Cited by
-
FOXM1: a new therapeutic target of extramammary Paget disease.Sci Rep. 2024 Feb 19;14(1):4048. doi: 10.1038/s41598-024-54773-8. Sci Rep. 2024. PMID: 38374400 Free PMC article.
-
Targeting NANOS1 in triple-negative breast cancer: synergistic effects of digoxin and PD-1 inhibitors in modulating the tumor immune microenvironment.Front Oncol. 2025 Jan 24;14:1536406. doi: 10.3389/fonc.2024.1536406. eCollection 2024. Front Oncol. 2025. PMID: 39927118 Free PMC article.
-
Impacts of vaping and marijuana use on airway health as determined by exhaled breath condensate (EBC).Respir Res. 2025 Feb 21;26(1):63. doi: 10.1186/s12931-025-03147-3. Respir Res. 2025. PMID: 39984952 Free PMC article.
-
Characterization of the In Vitro Cultured Ovarian Cells in the Asian Yellow Pond Turtle (Mauremys mutica).Biology (Basel). 2022 Sep 26;11(10):1404. doi: 10.3390/biology11101404. Biology (Basel). 2022. PMID: 36290308 Free PMC article.
-
Emerging Roles of NANOS RNA-Binding Proteins in Cancer.Int J Mol Sci. 2022 Aug 20;23(16):9408. doi: 10.3390/ijms23169408. Int J Mol Sci. 2022. PMID: 36012673 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

