Protease and DNase Activities of a Very Stable High-Molecular-Mass Multiprotein Complex from Sea Cucumber Eupentacta fraudatrix
- PMID: 35743119
- PMCID: PMC9224385
- DOI: 10.3390/ijms23126677
Protease and DNase Activities of a Very Stable High-Molecular-Mass Multiprotein Complex from Sea Cucumber Eupentacta fraudatrix
Abstract
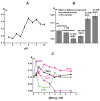
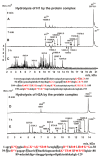
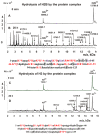
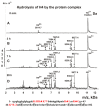
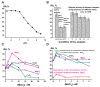
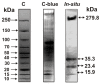
Only some human organs, including the liver, are capable of very weak self-regeneration. Some marine echinoderms are very useful for studying the self-regeneration processes of organs and tissues. For example, sea cucumbers Eupentacta fraudatrix (holothurians) demonstrate complete restoration of all organs and the body within several weeks after their division into two parts. Therefore, these cucumbers are a prospective model for studying the general mechanisms of self-regeneration. However, there is no data available yet concerning biomolecules of holothurians, which can stimulate the processes of organ and whole-body regeneration. Investigation of these restoration mechanisms is very important for modern medicine and biology because it can help to understand which hormones, nucleic acids, proteins, enzymes, or complexes play an essential role in self-regeneration. It is possible that stable, polyfunctional, high-molecular-weight protein complexes play an essential role in these processes. It has recently been shown that sea cucumbers Eupentacta fraudatrix contain a very stable multiprotein complex of about 2000 kDa. The first analysis of possible enzymatic activities of a stable protein complex was carried out in this work, revealing that the complex possesses several protease and DNase activities. The complex metalloprotease is activated by several metal ions (Zn2+ > Mn2+ > Mg2+). The relative contribution of metalloproteases (~63.4%), serine-like protease (~30.5%), and thiol protease (~6.1%) to the total protease activity of the complex was estimated. Metal-independent proteases of the complex hydrolyze proteins at trypsin-specific sites (after Lys and Arg). The complex contains both metal-dependent and metal-independent DNases. Mg2+, Mn2+, and Co2+ ions were found to strongly increase the DNase activity of the complex.
Keywords: protease and DNase catalytic activities of the complex; sea cucumber Eupentacta fraudatrix; very stable 2000 kDa protein complex.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Very Stable Two Mega Dalton High-Molecular-Mass Multiprotein Complex from Sea Cucumber Eupentacta fraudatrix.Molecules. 2021 Sep 21;26(18):5703. doi: 10.3390/molecules26185703. Molecules. 2021. PMID: 34577173 Free PMC article.
-
Comparison of the Content of Several Elements in Seawater, Sea Cucumber Eupentacta fraudatrix and Its High-Molecular-Mass Multiprotein Complex.Molecules. 2022 Mar 17;27(6):1958. doi: 10.3390/molecules27061958. Molecules. 2022. PMID: 35335321 Free PMC article.
-
Proteases from the regenerating gut of the holothurian Eupentacta fraudatrix.PLoS One. 2013;8(3):e58433. doi: 10.1371/journal.pone.0058433. Epub 2013 Mar 7. PLoS One. 2013. PMID: 23505505 Free PMC article.
-
DNases and apoptosis.Biochem Cell Biol. 2000;78(4):405-14. Biochem Cell Biol. 2000. PMID: 11012079 Review.
-
Molecular Aspects of Regeneration Mechanisms in Holothurians.Genes (Basel). 2021 Feb 10;12(2):250. doi: 10.3390/genes12020250. Genes (Basel). 2021. PMID: 33578707 Free PMC article. Review.
Cited by
-
Isolation of Extracellular Vesicles of Holothuria (Sea Cucumber Eupentacta fraudatrix).Int J Mol Sci. 2023 Aug 17;24(16):12907. doi: 10.3390/ijms241612907. Int J Mol Sci. 2023. PMID: 37629088 Free PMC article.
References
-
- Hyman L.H. The Invertebrates: Echinodermata. The Coelome Bilateria. McGraw-Hill Book Co.; New York, NY, USA: 1955. p. 763.
-
- Dolmatov I.Y., Mashanov V.S. Regeneration in Holothurians. Dalnauka; Vladivostok, Russia: 2007. pp. 1–212.
-
- Dolmatov I.Y. Regeneration of the digestive system in holothurians. Zh. Obshch. Biol. 2009;70:319–330. - PubMed
-
- Dolmatov I.Y. Regeneration of the aquapharyngeal complex in the holothurian Eupentacta fraudatrix (Holothuroidea, Dendrochirota) In: Taban C.H., Boilly B., editors. Keys for Regeneration. Volume 23. Karger; Basel, Germany: 1992. pp. 40–50. - PubMed
-
- Levin V.S., Dalnevostochnyi Trepang Biologiya, Dobycha, Vosproizvodstvo (The Far East Trepang Biology, Catching, Reproduction); St. Petersburg, Goland, Russia. 2000. [(accessed on 18 May 2022)]. Available online: https://search.rsl.ru/ru/record/01000663732.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

