Prediction and Experimental Validation of a New Salinity-Responsive Cis-Regulatory Element (CRE) in a Tilapia Cell Line
- PMID: 35743818
- PMCID: PMC9225295
- DOI: 10.3390/life12060787
Prediction and Experimental Validation of a New Salinity-Responsive Cis-Regulatory Element (CRE) in a Tilapia Cell Line
Abstract
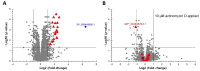
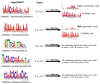
Transcriptional regulation is a major mechanism by which organisms integrate gene x environment interactions. It can be achieved by coordinated interplay between cis-regulatory elements (CREs) and transcription factors (TFs). Euryhaline tilapia (Oreochromis mossambicus) tolerate a wide range of salinity and thus are an appropriate model to examine transcriptional regulatory mechanisms during salinity stress in fish. Quantitative proteomics in combination with the transcription inhibitor actinomycin D revealed 19 proteins that are transcriptionally upregulated by hyperosmolality in tilapia brain (OmB) cells. We searched the extended proximal promoter up to intron1 of each corresponding gene for common motifs using motif discovery tools. The top-ranked motif identified (STREME1) represents a binding site for the Forkhead box TF L1 (FoxL1). STREME1 function during hyperosmolality was experimentally validated by choosing two of the 19 genes, chloride intracellular channel 2 (clic2) and uridine phosphorylase 1 (upp1), that are enriched in STREME1 in their extended promoters. Transcriptional induction of these genes during hyperosmolality requires STREME1, as evidenced by motif mutagenesis. We conclude that STREME1 represents a new functional CRE that contributes to gene x environment interactions during salinity stress in tilapia. Moreover, our results indicate that FoxL1 family TFs are contribute to hyperosmotic induction of genes in euryhaline fish.
Keywords: cellular osmoregulation; cis-regulatory element; fish; salinity; stress tolerance; transcription factor; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
An osmolality/salinity-responsive enhancer 1 (OSRE1) in intron 1 promotes salinity induction of tilapia glutamine synthetase.Sci Rep. 2020 Jul 21;10(1):12103. doi: 10.1038/s41598-020-69090-z. Sci Rep. 2020. PMID: 32694739 Free PMC article.
-
Osmolality/salinity-responsive enhancers (OSREs) control induction of osmoprotective genes in euryhaline fish.Proc Natl Acad Sci U S A. 2017 Mar 28;114(13):E2729-E2738. doi: 10.1073/pnas.1614712114. Epub 2017 Mar 13. Proc Natl Acad Sci U S A. 2017. PMID: 28289196 Free PMC article.
-
Transcriptional upregulation of the myo-inositol biosynthesis pathway is enhanced by NFAT5 in hyperosmotically stressed tilapia cells.Am J Physiol Cell Physiol. 2024 Sep 1;327(3):C545-C556. doi: 10.1152/ajpcell.00187.2024. Epub 2024 Jul 1. Am J Physiol Cell Physiol. 2024. PMID: 38946247
-
Transcriptional regulation of prolactin in a euryhaline teleost: Characterisation of gene promoters through in silico and transcriptome analyses.J Neuroendocrinol. 2020 Nov;32(11):e12905. doi: 10.1111/jne.12905. Epub 2020 Sep 29. J Neuroendocrinol. 2020. PMID: 32996203 Free PMC article. Review.
-
Endocrine and osmoregulatory responses to tidally-changing salinities in fishes.Gen Comp Endocrinol. 2022 Sep 15;326:114071. doi: 10.1016/j.ygcen.2022.114071. Epub 2022 Jun 11. Gen Comp Endocrinol. 2022. PMID: 35697315 Review.
Cited by
-
Removal of evolutionarily conserved functional MYC domains in a tilapia cell line using a vector-based CRISPR/Cas9 system.Sci Rep. 2023 Jul 26;13(1):12086. doi: 10.1038/s41598-023-37928-x. Sci Rep. 2023. PMID: 37495710 Free PMC article.
References
-
- Cooper G.M. The Cell: A Molecular Approach. 2nd ed. Sinauer Associates; Sunderland, MA, USA: 2000. Regulation of Transcription in Eukaryotes.
Grants and funding
- IOS-1656371/National Science Foundation
- IS-4800-15 R/United States-Israel Binational Agricultural Research and Development Fund
- CA-D-ASC-7690-H/California Agricultural Experiment Station
- CA-D-ASC-7624/California Agricultural Experiment Station
- CA-D-ASC-2667-RR/California Agricultural Experiment Station
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

