Culturable Bacterial Diversity from the Basaltic Subsurface of the Young Volcanic Island of Surtsey, Iceland
- PMID: 35744695
- PMCID: PMC9229223
- DOI: 10.3390/microorganisms10061177
Culturable Bacterial Diversity from the Basaltic Subsurface of the Young Volcanic Island of Surtsey, Iceland
Abstract
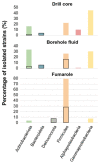
The oceanic crust is the world's largest and least explored biosphere on Earth. The basaltic subsurface of Surtsey island in Iceland represents an analog of the warm and newly formed-oceanic crust and offers a great opportunity for discovering novel microorganisms. In this study, we collected borehole fluids, drill cores, and fumarole samples to evaluate the culturable bacterial diversity from the subsurface of the island. Enrichment cultures were performed using different conditions, media and temperatures. A total of 195 bacterial isolates were successfully cultivated, purified, and identified based on MALDI-TOF MS analysis and by 16S rRNA gene sequencing. Six different clades belonging to Firmicutes (40%), Gammaproteobacteria (28.7%), Actinobacteriota (22%), Bacteroidota (4.1%), Alphaproteobacteria (3%), and Deinococcota (2%) were identified. Bacillus (13.3%) was the major genus, followed by Geobacillus (12.33%), Enterobacter (9.23%), Pseudomonas (6.15%), and Halomonas (5.64%). More than 13% of the cultured strains potentially represent novel species based on partial 16S rRNA gene sequences. Phylogenetic analyses revealed that the isolated strains were closely related to species previously detected in soil, seawater, and hydrothermal active sites. The 16S rRNA gene sequences of the strains were aligned against Amplicon Sequence Variants (ASVs) from the previously published 16S rRNA gene amplicon sequence datasets obtained from the same samples. Compared with the culture-independent community composition, only 5 out of 49 phyla were cultivated. However, those five phyla accounted for more than 80% of the ASVs. Only 121 out of a total of 5642 distinct ASVs were culturable (≥98.65% sequence similarity), representing less than 2.15% of the ASVs detected in the amplicon dataset. Here, we support that the subsurface of Surtsey volcano hosts diverse and active microbial communities and that both culture-dependent and -independent methods are essential to improving our insight into such an extreme and complex volcanic environment.
Keywords: Iceland; Surtsey; bacteria; culturable microbial diversity; extreme environment; oceanic subsurface.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Magnabosco C., Lin L.H., Dong H., Bomberg M., Ghiorse W., Stan-Lotter H., Pedersen K., Kieft T.L., van Heerden E., Onstott T.C. The Biomass and Biodiversity of the Continental Subsurface. Nat. Geosci. 2018;1:707–717. doi: 10.1038/s41561-018-0221-6. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

