Resistance Is Not Futile: The Role of Quorum Sensing Plasticity in Pseudomonas aeruginosa Infections and Its Link to Intrinsic Mechanisms of Antibiotic Resistance
- PMID: 35744765
- PMCID: PMC9228389
- DOI: 10.3390/microorganisms10061247
Resistance Is Not Futile: The Role of Quorum Sensing Plasticity in Pseudomonas aeruginosa Infections and Its Link to Intrinsic Mechanisms of Antibiotic Resistance
Abstract
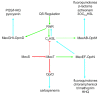
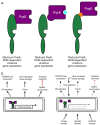
Bacteria use a cell-cell communication process called quorum sensing (QS) to orchestrate collective behaviors. QS relies on the group-wide detection of extracellular signal molecules called autoinducers (AI). Quorum sensing is required for virulence and biofilm formation in the human pathogen Pseudomonas aeruginosa. In P. aeruginosa, LasR and RhlR are homologous LuxR-type soluble transcription factor receptors that bind their cognate AIs and activate the expression of genes encoding functions required for virulence and biofilm formation. While some bacterial signal transduction pathways follow a linear circuit, as phosphoryl groups are passed from one carrier protein to another ultimately resulting in up- or down-regulation of target genes, the QS system in P. aeruginosa is a dense network of receptors and regulators with interconnecting regulatory systems and outputs. Once activated, it is not understood how LasR and RhlR establish their signaling hierarchy, nor is it clear how these pathway connections are regulated, resulting in chronic infection. Here, we reviewed the mechanisms of QS progression as it relates to bacterial pathogenesis and antimicrobial resistance and tolerance.
Keywords: antibiotic resistance; quorum sensing; virulence.
Conflict of interest statement
The authors declare that they have no conflict of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Figures

References
-
- Redfield R.R. Antibiotic Resistance Threats in the United States. Centres for Disease Control and Prevention; Atlanta, GA, USA: 2019.
-
- Centers for Disease Control and Prevention . Biggest Threats and Data: 2019 AR Threats Report. CDC; Atlanta, GA, USA: 2019.
-
- Bjarnsholt T., Jensen P.O., Jakobsen T.H., Phipps R., Nielsen A.K., Rybtke M.T., Tolker-Nielsen T., Givskov M., Hoiby N., Ciofu O., et al. Quorum sensing and virulence of Pseudomonas aeruginosa during lung infection of cystic fibrosis patients. PLoS ONE. 2010;5:e10115. doi: 10.1371/journal.pone.0010115. - DOI - PMC - PubMed
-
- Wolter D.J., Lister P.D. Mechanisms of beta-lactam resistance among Pseudomonas aeruginosa. [(accessed on 25 November 2021)];Curr. Pharm. Des. 2013 19:209–222. doi: 10.2174/138161213804070311. Available online: https://www.ncbi.nlm.nih.gov/pubmed/22894618. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

