Synthesis and Activity of Triazole-Adenosine Analogs as Protein Arginine Methyltransferase 5 Inhibitors
- PMID: 35744905
- PMCID: PMC9228412
- DOI: 10.3390/molecules27123779
Synthesis and Activity of Triazole-Adenosine Analogs as Protein Arginine Methyltransferase 5 Inhibitors
Abstract
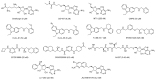
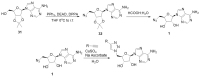
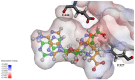
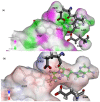
Protein arginine methyltransferase 5 (PRMT5) is an attractive molecular target in anticancer drug discovery due to its extensive involvement in transcriptional control, RNA processing, and other cellular pathways that are causally related to tumor initiation and progression. In recent years, various compounds have been screened or designed to target either the substrate- or cofactor-binding site of PRMT5. To expand the diversity of chemotypes for inhibitory binding to PRMT5 and other AdoMet-dependent methyltransferases, in this work, we designed a series of triazole-containing adenosine analogs aimed at targeting the cofactor-binding site of PRMT5. Triazole rings have commonly been utilized in drug discovery due to their ease of synthesis and functionalization as bioisosteres of amide bonds. Herein, we utilized the electronic properties of the triazole ring as a novel way to specifically target the cofactor-binding site of PRMT5. A total of about 30 compounds were synthesized using the modular alkyne-azide cycloaddition reaction. Biochemical tests showed that these compounds exhibited inhibitory activity of PRMT5 at varying degrees and several showed single micromolar potency, with clear selectivity for PRMT5 over PRMT1. Docking-based structural analysis showed that the triazole ring plays a key role in binding to the characteristic residue Phe327 in the active pocket of PRMT5, explaining the compounds' selectivity for this type-II enzyme. Overall, this work provides new structure-activity relationship information on the design of AdoMet analogs for selective inhibition of PRMT5. Further structural optimization work will further improve the potency of the top leads.
Keywords: PRMT5; SAM analog; arginine methylation; click chemistry; epigenetics; inhibitor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Rational Design, synthesis and biological evaluation of novel triazole derivatives as potent and selective PRMT5 inhibitors with antitumor activity.J Comput Aided Mol Des. 2019 Aug;33(8):775-785. doi: 10.1007/s10822-019-00214-y. Epub 2019 Jul 16. J Comput Aided Mol Des. 2019. PMID: 31312965
-
Discovery of new potent protein arginine methyltransferase 5 (PRMT5) inhibitors by assembly of key pharmacophores from known inhibitors.Bioorg Med Chem Lett. 2018 Dec 15;28(23-24):3693-3699. doi: 10.1016/j.bmcl.2018.10.026. Epub 2018 Oct 19. Bioorg Med Chem Lett. 2018. PMID: 30366617
-
Discovery of Novel PRMT5 Inhibitors by Virtual Screening and Biological Evaluations.Chem Pharm Bull (Tokyo). 2019;67(4):382-388. doi: 10.1248/cpb.c18-00980. Chem Pharm Bull (Tokyo). 2019. PMID: 30930442
-
Nucleoside protein arginine methyltransferase 5 (PRMT5) inhibitors.Bioorg Med Chem Lett. 2019 Jun 1;29(11):1264-1269. doi: 10.1016/j.bmcl.2019.03.042. Epub 2019 Mar 27. Bioorg Med Chem Lett. 2019. PMID: 30956011 Review.
-
Targeting protein arginine methyltransferase 5 in disease.Future Med Chem. 2017 Nov;9(17):2081-2098. doi: 10.4155/fmc-2017-0089. Epub 2017 Oct 27. Future Med Chem. 2017. PMID: 29076773 Review.
Cited by
-
Recent updates in click and computational chemistry for drug discovery and development.Front Chem. 2023 Feb 7;11:1114970. doi: 10.3389/fchem.2023.1114970. eCollection 2023. Front Chem. 2023. PMID: 36825226 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

