Catalytic Properties of Caseinolytic Protease Subunit of Plasmodium knowlesi and Its Inhibition by a Member of δ-Lactone, Hyptolide
- PMID: 35744912
- PMCID: PMC9228282
- DOI: 10.3390/molecules27123787
Catalytic Properties of Caseinolytic Protease Subunit of Plasmodium knowlesi and Its Inhibition by a Member of δ-Lactone, Hyptolide
Abstract
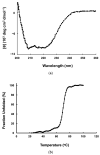
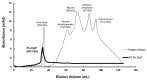
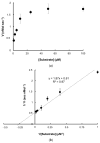
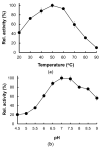
The caseinolytic protease (Clp) system plays an essential role in the protein homeostasis of the malaria parasite, particularly at the stage of apicoplast development. The inhibition of this protein is known to have a lethal effect on the parasite and is therefore considered an interesting avenue for antimalaria drugs discovery. The catalytic activity of the Clp system is modulated by its proteolytic subunit (ClpP), which belongs to the serine protease family member and is therefore extensively studied for further inhibitors development. Among many inhibitors, the group of β-lactone is known to be a specific inhibitor for ClpP. Nevertheless, other groups of lactones have never been studied. This study aims to characterize the catalytic properties of ClpP of Plasmodium knowlesi (Pk-ClpP) and the inhibition properties of a δ-lactone hyptolide against this protein. Accordingly, a codon-optimized synthetic gene encoding Pk-ClpP was expressed in Escherichia coli BL21(DE3) and purified under a single step of Ni2+-affinity chromatography, yielding a 2.20 mg from 1 L culture. Meanwhile, size-exclusion chromatography indicated that Pk-ClpP migrated primarily as homoheptameric with a size of 205 kDa. The specific activity of pure Pk-ClpP was 0.73 U µg-1, with a catalytic efficiency kcat/KM of 0.05 µM-1 s-1, with optimum temperature and pH of 50 °C and 7.0-7.5, respectively. Interestingly, hyptolide, a member of δ-lactone, was shown to inhibit Pk-ClpP with an IC50 value of 17.36 ± 1.44 nM. Structural homology modelling, secondary structure prediction, and far-UV CD spectra revealed that helical structures dominate this protein. In addition, the structural homology modeling showed that this protein forms a barrel-shaped homoheptamer. Docking simulation revealed that the inhibition was found to be a competitive inhibition in which hyptolide was able to dock into the catalytic site and block the substrate. The competitiveness of hyptolide is due to the higher binding affinity of this molecule than the substrate.
Keywords: Plasmodium knowlesi; antimalarial drug; caseinolytic protease; malaria; δ-lactone.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Technical data on the inhibition properties of some medicinal plant extracts towards caseinolytic protease proteolytic subunit of Plasmodium knowlesi.Data Brief. 2021 Nov 20;39:107588. doi: 10.1016/j.dib.2021.107588. eCollection 2021 Dec. Data Brief. 2021. PMID: 34877373 Free PMC article.
-
Phenyl Esters Are Potent Inhibitors of Caseinolytic Protease P and Reveal a Stereogenic Switch for Deoligomerization.J Am Chem Soc. 2015 Jul 8;137(26):8475-83. doi: 10.1021/jacs.5b03084. Epub 2015 Jun 29. J Am Chem Soc. 2015. PMID: 26083639
-
Structure of Staphylococcus aureus ClpP Bound to the Covalent Active-Site Inhibitor Cystargolide A.Angew Chem Int Ed Engl. 2024 Jan 15;63(3):e202314028. doi: 10.1002/anie.202314028. Epub 2023 Dec 12. Angew Chem Int Ed Engl. 2024. PMID: 38029352
-
Dynamics of the ClpP serine protease: a model for self-compartmentalized proteases.Crit Rev Biochem Mol Biol. 2014 Sep-Oct;49(5):400-12. doi: 10.3109/10409238.2014.925421. Epub 2014 Jun 10. Crit Rev Biochem Mol Biol. 2014. PMID: 24915503 Review.
-
The development of small-molecule modulators for ClpP protease activity.Mol Biosyst. 2016 Dec 20;13(1):23-31. doi: 10.1039/c6mb00644b. Mol Biosyst. 2016. PMID: 27831584 Review.
Cited by
-
The Establishment and Application of Indirect 3AB-ELISA for the Detection of Antibodies against Senecavirus A.Viruses. 2023 Mar 28;15(4):861. doi: 10.3390/v15040861. Viruses. 2023. PMID: 37112841 Free PMC article.
-
Purification and characterization of cysteine protease of Sarcocystis fusiformis from infected Egyptian water buffaloes.Sci Rep. 2023 Sep 26;13(1):16123. doi: 10.1038/s41598-023-43147-1. Sci Rep. 2023. PMID: 37752241 Free PMC article.
References
-
- Kotepui M., Kotepui K.U., Milanez G.D.J., Masangkay F.R. Prevalence of severe Plasmodium knowlesi infection and risk factors related to severe complications compared with non-severe P. knowlesi and severe P. falciparum malaria: A systematic review and meta-analysis. Infect. Dis. Poverty. 2020;9:106. doi: 10.1186/s40249-020-00727-x. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

