Recent Advances in Rapid and Highly Sensitive Detection of Proteins and Specific DNA Sequences Using a Magnetic Modulation Biosensing System
- PMID: 35746278
- PMCID: PMC9230956
- DOI: 10.3390/s22124497
Recent Advances in Rapid and Highly Sensitive Detection of Proteins and Specific DNA Sequences Using a Magnetic Modulation Biosensing System
Abstract
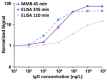
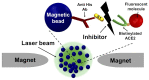
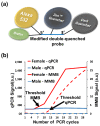
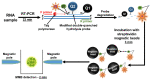
In early disease stages, biomolecules of interest exist in very low concentrations, presenting a significant challenge for analytical devices and methods. Here, we provide a comprehensive overview of an innovative optical biosensing technology, termed magnetic modulation biosensing (MMB), its biomedical applications, and its ongoing development. In MMB, magnetic beads are attached to fluorescently labeled target molecules. A controlled magnetic force aggregates the magnetic beads and transports them in and out of an excitation laser beam, generating a periodic fluorescent signal that is detected and demodulated. MMB applications include rapid and highly sensitive detection of specific nucleic acid sequences, antibodies, proteins, and protein interactions. Compared with other established analytical methodologies, MMB provides improved sensitivity, shorter processing time, and simpler protocols.
Keywords: biosensing; magnetic beads; magnetic modulation biosensing; nucleic acids; protein-protein interactions; serology.
Conflict of interest statement
Amos Danielli has a financial interest in MagBiosense, Inc., which, however, did not financially support this work.
Figures








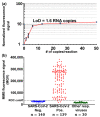
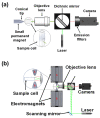


References
-
- Rissin D.M., Kan C.W., Campbell T.G., Howes S.C., Fournier D.R., Song L., Piech T., Patel P.P., Chang L., Rivnak A.J., et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotech. 2010;28:595–599. doi: 10.1038/nbt.1641. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

