Epitope-Based Vaccines against the Chlamydia trachomatis Major Outer Membrane Protein Variable Domain 4 Elicit Protection in Mice
- PMID: 35746483
- PMCID: PMC9227494
- DOI: 10.3390/vaccines10060875
Epitope-Based Vaccines against the Chlamydia trachomatis Major Outer Membrane Protein Variable Domain 4 Elicit Protection in Mice
Abstract
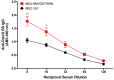
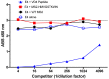
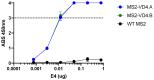
Chlamydia trachomatis (Ct) is the most common bacterial sexual transmitted pathogen, yet a vaccine is not currently available. Here, we used the immunogenic bacteriophage MS2 virus-like particle (VLP) technology to engineer vaccines against the Ct major outer membrane protein variable domain 4 (MOMP-VD4), which contains a conserved neutralizing epitope (TTLNPTIAG). A previously described monoclonal antibody to the MOMP-VD4 (E4 mAb) is capable of neutralizing all urogenital Ct serovars and binds this core epitope, as well as several non-contiguous amino acids. This suggests that this core epitope may require conformational context in order to elicit neutralizing antibodies to Ct. In order to identify immunogens that could elicit neutralizing antibodies to the TTLNPTIAG epitope, we used two approaches. First, we used affinity selection with a bacteriophage MS2-VLP library displaying random peptides in a constrained, surface-exposed loop to identify potential E4 mAb mimotopes. After four rounds of affinity selection, we identified a VLP-displayed peptide (HMVGSTKWTN) that could bind to the E4 mAb and elicited serum IgG that bound weakly to Ct elementary bodies by ELISA. Second, two versions of the core conserved TTLNPTIAG epitope (TTLNPTIAG and TTLNPTIAGA) were recombinantly expressed on the coat protein of the MS2 VLP in a constrained, surface-exposed loop. Mouse immune sera IgG bound to Ct elementary bodies by ELISA. Immunization with these MS2 VLPs provided protection from vaginal Chlamydia infection in a murine challenge model. These data suggest that short peptide epitopes targeting the MOMP-VD4 could be appropriate for Ct vaccine design when displayed on an immunogenic bacteriophage VLP vaccine platform.
Keywords: Chlamydia trachomatis; affinity selection; antibodies; bacteriophage; epitope; vaccine; virus-like particle.
Conflict of interest statement
A.L.C. and K.M.F. are inventors on a patent pending for vaccines described in this manuscript. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Gottlieb S.L., Deal C.D., Giersing B., Rees H., Bolan G., Johnston C., Timms P., Gray-Owen S.D., Jerse A.E., Cameron C.E., et al. The Global Roadmap for Advancing Development of Vaccines against Sexually Transmitted Infections: Update and next Steps. Vaccine. 2016;34:2939–2947. doi: 10.1016/j.vaccine.2016.03.111. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

