Adenovirus DNA Polymerase Loses Fidelity on a Stretch of Eleven Homocytidines during Pre-GMP Vaccine Preparation
- PMID: 35746566
- PMCID: PMC9227658
- DOI: 10.3390/vaccines10060960
Adenovirus DNA Polymerase Loses Fidelity on a Stretch of Eleven Homocytidines during Pre-GMP Vaccine Preparation
Abstract
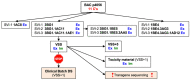
In this study, we invented and construct novel candidate HIV-1 vaccines. Through genetic and protein engineering, we unknowingly constructed an HIV-1-derived transgene with a homopolymeric run of 11 cytidines, which was inserted into an adenovirus vaccine vector. Here, we describe the virus rescue, three rounds of clonal purification and preparation of good manufacturing practise (GMP) starting material assessed for genetic stability in five additional virus passages. Throughout these steps, quality control assays indicated the presence of the transgene in the virus genome, expression of the correct transgene product and immunogenicity in mice. However, DNA sequencing of the transgene revealed additional cytidines inserted into the original 11-cytidine region, and the GMP manufacture had to be aborted. Subsequent analyses indicated that as little as 1/25th of the virus dose used for confirmation of protein expression (106 cells at a multiplicity of infection of 10) and murine immunogenicity (108 infectious units per animal) met the quality acceptance criteria. Similar frameshifts in the expressed proteins were reproduced in a one-reaction in vitro transcription/translation employing phage T7 polymerase and E. coli ribosomes. Thus, the most likely mechanism for addition of extra cytidines into the ChAdOx1.tHIVconsv6 genome is that the adenovirus DNA polymerase lost its fidelity on a stretch of 11 cytidines, which informs future adenovirus vaccine designs.
Keywords: HIVconsvX; T7 polymerase; adenovirus DNA polymerase; polymerase fidelity; protein engineering; vaccines.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Adenovirus Transcriptome in Human Cells Infected with ChAdOx1-Vectored Candidate HIV-1 Vaccine Is Dominated by High Levels of Correctly Spliced HIVconsv1&62 Transgene RNA.Vaccines (Basel). 2023 Jul 1;11(7):1187. doi: 10.3390/vaccines11071187. Vaccines (Basel). 2023. PMID: 37515003 Free PMC article.
-
Inclusion of the murine IgGκ signal peptide increases the cellular immunogenicity of a simian adenoviral vectored Plasmodium vivax multistage vaccine.Vaccine. 2018 May 11;36(20):2799-2808. doi: 10.1016/j.vaccine.2018.03.091. Epub 2018 Apr 12. Vaccine. 2018. PMID: 29657070 Free PMC article.
-
Enterovirus A71 Containing Codon-Deoptimized VP1 and High-Fidelity Polymerase as Next-Generation Vaccine Candidate.J Virol. 2019 Jun 14;93(13):e02308-18. doi: 10.1128/JVI.02308-18. Print 2019 Jul 1. J Virol. 2019. PMID: 30996087 Free PMC article.
-
Influenza virus vaccine live intranasal--MedImmune vaccines: CAIV-T, influenza vaccine live intranasal.Drugs R D. 2003;4(5):312-9. doi: 10.2165/00126839-200304050-00007. Drugs R D. 2003. PMID: 12952502 Review.
-
Adenovirus Vectors: Excellent Tools for Vaccine Development.Immune Netw. 2021 Feb 15;21(1):e6. doi: 10.4110/in.2021.21.e6. eCollection 2021 Feb. Immune Netw. 2021. PMID: 33728099 Free PMC article. Review.
Cited by
-
Design, Immunogenicity and Preclinical Efficacy of the ChAdOx1.COVconsv12 Pan-Sarbecovirus T-Cell Vaccine.Vaccines (Basel). 2024 Aug 26;12(9):965. doi: 10.3390/vaccines12090965. Vaccines (Basel). 2024. PMID: 39339997 Free PMC article.
-
Adenovirus Transcriptome in Human Cells Infected with ChAdOx1-Vectored Candidate HIV-1 Vaccine Is Dominated by High Levels of Correctly Spliced HIVconsv1&62 Transgene RNA.Vaccines (Basel). 2023 Jul 1;11(7):1187. doi: 10.3390/vaccines11071187. Vaccines (Basel). 2023. PMID: 37515003 Free PMC article.
References
-
- Plotkin S.A., Plotkin S. A Short History of Vaccination. 5th ed. Elsevier-Saunders; Philadelphia, PA, USA: 2008.
-
- Chong S.S.F., Kim M., Limoli M., Obscherning E., Wu P., Feisee L., Nakashima N., Lim J.C.W. Measuring progress of regulatory convergence and cooperation among Asia–Pacific Economic Cooperation (APEC) member economies in the context of the COVID-19 pandemic. Ther. Innov. Regul. Sci. 2021;55:786–798. doi: 10.1007/s43441-021-00285-w. - DOI - PMC - PubMed
Grants and funding
- HHSN27220110002II/HHSN27200037, Subcontract No.: OX-14007-004-0037-212/NH/NIH HHS/United States
- MR/N023668/1/MRC_/Medical Research Council/United Kingdom
- SRIA2015-1066/European & Developing Countries Clinical Trials Partnership
- 681137-EAVI2020/European Commission
- SOW5/International AIDS Vaccine Initiative
LinkOut - more resources
Full Text Sources
Miscellaneous

