Differential Pathogenesis of SARS-CoV-2 Variants of Concern in Human ACE2-Expressing Mice
- PMID: 35746611
- PMCID: PMC9231291
- DOI: 10.3390/v14061139
Differential Pathogenesis of SARS-CoV-2 Variants of Concern in Human ACE2-Expressing Mice
Abstract
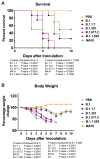
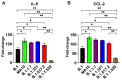
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the current pandemic, resulting in millions of deaths worldwide. Increasingly contagious variants of concern (VoC) have fueled recurring global infection waves. A major question is the relative severity of the disease caused by previous and currently circulating variants of SARS-CoV-2. In this study, we evaluated the pathogenesis of SARS-CoV-2 variants in human ACE-2-expressing (K18-hACE2) mice. Eight-week-old K18-hACE2 mice were inoculated intranasally with a representative virus from the original B.1 lineage or from the emerging B.1.1.7 (alpha), B.1.351 (beta), B.1.617.2 (delta), or B.1.1.529 (omicron) lineages. We also infected a group of mice with the mouse-adapted SARS-CoV-2 (MA10). Our results demonstrate that B.1.1.7, B.1.351 and B.1.617.2 viruses are significantly more lethal than the B.1 strain in K18-hACE2 mice. Infection with the B.1.1.7, B.1.351, and B.1.617.2 variants resulted in significantly higher virus titers in the lungs and brain of mice compared with the B.1 virus. Interestingly, mice infected with the B.1.1.529 variant exhibited less severe clinical signs and a high survival rate. We found that B.1.1.529 replication was significantly lower in the lungs and brain of infected mice in comparison with other VoC. The transcription levels of cytokines and chemokines in the lungs of B.1- and B.1.1.529-infected mice were significantly less when compared with those challenged with other VoC. Together, our data provide insights into the pathogenesis of previous and circulating SARS-CoV-2 VoC in mice.
Keywords: ACE2-expressing mice; COVID-19; SARS-CoV-2 variants; inflammation; omicron.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Khan A., Zia T., Suleman M., Khan T., Ali S.S., Abbasi A.A., Mohammad A., Wei D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J. Cell Physiol. 2021;236:7045–7057. doi: 10.1002/jcp.30367. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

