Memory CD8 T Cells Protect against Cytomegalovirus Disease by Formation of Nodular Inflammatory Foci Preventing Intra-Tissue Virus Spread
- PMID: 35746617
- PMCID: PMC9229300
- DOI: 10.3390/v14061145
Memory CD8 T Cells Protect against Cytomegalovirus Disease by Formation of Nodular Inflammatory Foci Preventing Intra-Tissue Virus Spread
Abstract
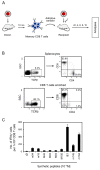
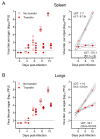
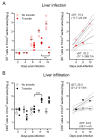
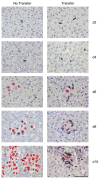
Cytomegaloviruses (CMVs) are controlled by innate and adaptive immune responses in an immunocompetent host while causing multiple organ diseases in an immunocompromised host. A risk group of high clinical relevance comprises transiently immunocompromised recipients of hematopoietic cell transplantation (HCT) in the "window of risk" between eradicative therapy of hematopoietic malignancies and complete reconstitution of the immune system. Cellular immunotherapy by adoptive transfer of CMV-specific CD8 T cells is an option to prevent CMV disease by controlling a primary or reactivated infection. While experimental models have revealed a viral epitope-specific antiviral function of cognate CD8 T cells, the site at which control is exerted remained unidentified. The observation that remarkably few transferred cells protect all organs may indicate an early blockade of virus dissemination from a primary site of productive infection to various target organs. Alternatively, it could indicate clonal expansion of a few transferred CD8 T cells for preventing intra-tissue virus spread after successful initial organ colonization. Our data in the mouse model of murine CMV infection provide evidence in support of the second hypothesis. We show that transferred cells vigorously proliferate to prevent virus spread, and thus viral histopathology, by confining and eventually resolving tissue infection within nodular inflammatory foci.
Keywords: adoptive cell transfer; antiviral protection; cytomegalovirus (CMV); growth kinetics; histopathology; immunotherapy; liver infection; memory CD8 T cells; nodular inflammatory focus (NIF); virus spread.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Immunotherapy of cytomegalovirus infection by low-dose adoptive transfer of antiviral CD8 T cells relies on substantial post-transfer expansion of central memory cells but not effector-memory cells.PLoS Pathog. 2023 Nov 16;19(11):e1011643. doi: 10.1371/journal.ppat.1011643. eCollection 2023 Nov. PLoS Pathog. 2023. PMID: 37972198 Free PMC article.
-
Therapeutic Vaccination of Hematopoietic Cell Transplantation Recipients Improves Protective CD8 T-Cell Immunotherapy of Cytomegalovirus Infection.Front Immunol. 2021 Aug 19;12:694588. doi: 10.3389/fimmu.2021.694588. eCollection 2021. Front Immunol. 2021. PMID: 34489940 Free PMC article.
-
Insufficient Antigen Presentation Due to Viral Immune Evasion Explains Lethal Cytomegalovirus Organ Disease After Allogeneic Hematopoietic Cell Transplantation.Front Cell Infect Microbiol. 2020 Apr 15;10:157. doi: 10.3389/fcimb.2020.00157. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32351904 Free PMC article.
-
Refining human T-cell immunotherapy of cytomegalovirus disease: a mouse model with 'humanized' antigen presentation as a new preclinical study tool.Med Microbiol Immunol. 2016 Dec;205(6):549-561. doi: 10.1007/s00430-016-0471-0. Epub 2016 Aug 18. Med Microbiol Immunol. 2016. PMID: 27539576 Review.
-
Cytomegalovirus immune evasion sets the functional avidity threshold for protection by CD8 T cells.Med Microbiol Immunol. 2023 Apr;212(2):153-163. doi: 10.1007/s00430-022-00733-w. Epub 2022 Apr 1. Med Microbiol Immunol. 2023. PMID: 35364731 Free PMC article. Review.
Cited by
-
Refractory cytomegalovirus infections in Chinese patients receiving allogeneic hematopoietic cell transplantation: a review of the literature.Front Immunol. 2023 Dec 22;14:1287456. doi: 10.3389/fimmu.2023.1287456. eCollection 2023. Front Immunol. 2023. PMID: 38187387 Free PMC article. Review.
-
Direct antigen presentation is the canonical pathway of cytomegalovirus CD8 T-cell priming regulated by balanced immune evasion ensuring a strong antiviral response.Front Immunol. 2023 Dec 12;14:1272166. doi: 10.3389/fimmu.2023.1272166. eCollection 2023. Front Immunol. 2023. PMID: 38149242 Free PMC article.
-
Immunotherapy of cytomegalovirus infection by low-dose adoptive transfer of antiviral CD8 T cells relies on substantial post-transfer expansion of central memory cells but not effector-memory cells.PLoS Pathog. 2023 Nov 16;19(11):e1011643. doi: 10.1371/journal.ppat.1011643. eCollection 2023 Nov. PLoS Pathog. 2023. PMID: 37972198 Free PMC article.
-
Immune-checkpoint expression in antigen-presenting cells (APCs) of cytomegaloviruses infection after transplantation: as a diagnostic biomarker.Arch Microbiol. 2023 Jul 10;205(8):280. doi: 10.1007/s00203-023-03623-8. Arch Microbiol. 2023. PMID: 37430000 Review.
-
Cytomegalovirus inhibitors of programmed cell death restrict antigen cross-presentation in the priming of antiviral CD8 T cells.PLoS Pathog. 2024 Aug 15;20(8):e1012173. doi: 10.1371/journal.ppat.1012173. eCollection 2024 Aug. PLoS Pathog. 2024. PMID: 39146364 Free PMC article.
References
-
- Davison A.J., Holton M., Dolan A., Dargan D.J., Gatherer D., Hayward G.S. Comparative genomics of primate cytomegaloviruses. In: Reddehase M.J., editor. Cytomegaloviruses: From Molecular Pathogenesis to Intervention. Volume 1. Caister Academic Press; Norfolk, UK: 2013. pp. 1–22.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

