Identification and Characterization of Two Novel Noda-like Viruses from Rice Plants Showing the Dwarfing Symptom
- PMID: 35746632
- PMCID: PMC9231309
- DOI: 10.3390/v14061159
Identification and Characterization of Two Novel Noda-like Viruses from Rice Plants Showing the Dwarfing Symptom
Abstract
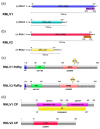
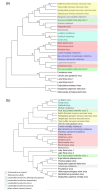
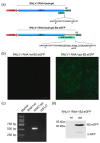
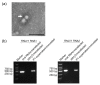
Nodaviruses are small bipartite RNA viruses and are considered animal viruses. Here, we identified two novel noda-like viruses (referred to as rice-associated noda-like virus 1 (RNLV1) and rice-associated noda-like virus 2 (RNLV2)) in field-collected rice plants showing a dwarfing phenotype through RNA-seq. RNLV1 genome consists of 3335 nt RNA1 and 1769 nt RNA2, and RNLV2 genome consists of 3279 nt RNA1 and 1525 nt RNA2. Three conserved ORFs were identified in each genome of the two novel viruses, encoding an RNA-dependent RNA polymerase, an RNA silencing suppressor, and a capsid protein, respectively. The results of sequence alignment, protein domain prediction, and evolutionary analysis indicate that these two novel viruses are clearly different from the known nodaviruses, especially the CPs. We have also determined that the B2 protein encoded by the two new noda-like viruses can suppress RNA silencing in plants. Two reverse genetic systems were constructed and used to show that RNLV1 RNA1 can replicate in plant cells and RNLV1 can replicate in insect Sf9 cells. We have also found two unusual peptidase family A21 domains in the RNLV1 CP, and RNLV1 CP can self-cleave in acidic environments. These findings provide new knowledge of novel nodaviruses.
Keywords: RNA silencing suppressor; noda-like virus; nodavirus; reverse genetic system; rice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Characterization of burdock mottle virus, a novel member of the genus Benyvirus, and the identification of benyvirus-related sequences in the plant and insect genomes.Virus Res. 2013 Oct;177(1):75-86. doi: 10.1016/j.virusres.2013.07.015. Epub 2013 Jul 30. Virus Res. 2013. PMID: 23911632
-
Comparisons among the larger genome segments of six nodaviruses and their encoded RNA replicases.J Gen Virol. 2001 Aug;82(Pt 8):1855-1866. doi: 10.1099/0022-1317-82-8-1855. J Gen Virol. 2001. PMID: 11457991
-
Discovery and Genomic Function of a Novel Rice Dwarf-Associated Bunya-like Virus.Viruses. 2022 May 29;14(6):1183. doi: 10.3390/v14061183. Viruses. 2022. PMID: 35746655 Free PMC article.
-
A novel virus of the genus Cilevirus causing symptoms similar to citrus leprosis.Phytopathology. 2013 May;103(5):488-500. doi: 10.1094/PHYTO-07-12-0177-R. Phytopathology. 2013. PMID: 23268581
-
Comparative studies of T = 3 and T = 4 icosahedral RNA insect viruses.Arch Virol Suppl. 1994;9:497-512. doi: 10.1007/978-3-7091-9326-6_48. Arch Virol Suppl. 1994. PMID: 8032278 Free PMC article. Review.
Cited by
-
The Virome of Cocoa Fermentation-Associated Microorganisms.Viruses. 2024 Jul 31;16(8):1226. doi: 10.3390/v16081226. Viruses. 2024. PMID: 39205200 Free PMC article.
-
Discovery and characterization of a novel carlavirus in Ligularia jaluensis plants.Virol Sin. 2025 Feb;40(1):71-79. doi: 10.1016/j.virs.2024.11.003. Epub 2024 Nov 12. Virol Sin. 2025. PMID: 39542218 Free PMC article.
-
The State of the Art of Plant Virus Research in China.Viruses. 2024 Oct 21;16(10):1639. doi: 10.3390/v16101639. Viruses. 2024. PMID: 39459971 Free PMC article.
-
A novel varicosavirus associated with Orychophragmus violaceus in China.Arch Virol. 2025 May 7;170(6):121. doi: 10.1007/s00705-025-06283-9. Arch Virol. 2025. PMID: 40329141
-
The Identification of Viral Pathogens in a Physostegia virginiana Plant Using High-Throughput RNA Sequencing.Viruses. 2023 Sep 21;15(9):1972. doi: 10.3390/v15091972. Viruses. 2023. PMID: 37766378 Free PMC article.
References
-
- Longworth J.F., Archibald R.D. A virus of black beetle, Heteronychus arator (F.) (Coleoptera: Scarabaeidae) N. Z. J. Zool. 1975;2:233–236. doi: 10.1080/03014223.1975.9517874. - DOI
-
- Dearing S.C., Scotti P.D., Wigley P.J., Dhana S.D. A small RNA virus isolated from the grass grub, Costelytra zealandica (Coleoptera: Scarabaeidae) N. Z. J. Zool. 1980;7:267–269. doi: 10.1080/03014223.1980.10423785. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

