Non-Productive Infection of Glial Cells with SARS-CoV-2 in Hamster Organotypic Cerebellar Slice Cultures
- PMID: 35746689
- PMCID: PMC9227386
- DOI: 10.3390/v14061218
Non-Productive Infection of Glial Cells with SARS-CoV-2 in Hamster Organotypic Cerebellar Slice Cultures
Abstract
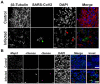
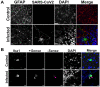
The numerous neurological syndromes associated with COVID-19 implicate an effect of viral pathogenesis on neuronal function, yet reports of direct SARS-CoV-2 infection in the brain are conflicting. We used a well-established organotypic brain slice culture to determine the permissivity of hamster brain tissues to SARS-CoV-2 infection. We found levels of live virus waned after inoculation and observed no evidence of cell-to-cell spread, indicating that SARS-CoV-2 infection was non-productive. Nonetheless, we identified a small number of infected cells with glial phenotypes; however, no evidence of viral infection or replication was observed in neurons. Our data corroborate several clinical studies that have assessed patients with COVID-19 and their association with neurological involvement.
Keywords: COVID-19; SARS-CoV-2; astrocytes; brain; microglia; neuroinflammation; organotypic culture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Beghi E., Michael M.B.D., Msc E.W., Winkler A.S., Global COVID-19 Neuro Research Coalition. Michael B.D., Solomon T., Westenberg E., Knauss S., Sejvar J., et al. Approaches to understanding COVID-19 and its neurological associations. Ann. Neurol. 2021;89:1059–1067. doi: 10.1002/ana.26076. - DOI - PMC - PubMed
-
- Raman B., Cassar M.P., Tunnicliffe E.M., Filippini N., Griffanti L., Alfaro-Almagro F., Okell T., Sheerin F., Xie C., Mahmod M., et al. Medium-term effects of SARS-CoV-2 infection on multiple vital organs, exercise capacity, cognition, quality of life and mental health, post-hospital discharge. eClinicalMedicine. 2021;31:100683. doi: 10.1016/j.eclinm.2020.100683. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

