Development of a Singleplex Real-Time Reverse Transcriptase PCR Assay for Pan-Dengue Virus Detection and Quantification
- PMID: 35746742
- PMCID: PMC9231192
- DOI: 10.3390/v14061271
Development of a Singleplex Real-Time Reverse Transcriptase PCR Assay for Pan-Dengue Virus Detection and Quantification
Abstract
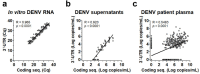
Dengue virus (DENV) infection is a significant global health problem. There are no specific therapeutics or widely available vaccines. Early diagnosis is critical for patient management. Viral RNA detection by multiplex RT-PCR using multiple pairs of primers/probes allowing the simultaneous detection of all four DENV serotypes is commonly used. However, increasing the number of primers in the RT-PCR reaction reduces the sensitivity of detection due to the increased possibility of primer dimer formation. Here, a one tube, singleplex real-time RT-PCR specific to DENV 3'-UTR was developed for the detection and quantification of pan-DENV with no cross reactivity to other flaviviruses. The sensitivity of DENV detection was as high as 96.9% in clinical specimens collected at the first day of hospitalization. Our assay provided equivalent PCR efficiency and RNA quantification among each DENV serotype. The assay's performance was comparable with previously established real-time RT-PCR targeting coding sequences. Using both assays on the same specimens, our results indicate the presence of defective virus particles in the circulation of patients infected with all serotypes. Dual regions targeting RT-PCR enhanced the sensitivity of viral genome detection especially during the late acute phase when viremia rapidly decline and an incomplete viral genome was clinically evident.
Keywords: DENV 3′-UTR detection; defective virus particles; dengue viral load quantification; dengue virus detection; dual regions detection; incomplete viral genome; one tube; singleplex real-time RT-PCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Disease G.B.D., Injury I., Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2018;392:1789–1858. doi: 10.1016/S0140-6736(18)32279-7. - DOI - PMC - PubMed
-
- Dengue Haemorrhagic Fever: Diagnosis, Treatment, Prevention and Control. World Health Organization; Geneva, Switzerland: 1997.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

