Targeted Virome Sequencing Enhances Unbiased Detection and Genome Assembly of Known and Emerging Viruses-The Example of SARS-CoV-2
- PMID: 35746743
- PMCID: PMC9227943
- DOI: 10.3390/v14061272
Targeted Virome Sequencing Enhances Unbiased Detection and Genome Assembly of Known and Emerging Viruses-The Example of SARS-CoV-2
Abstract
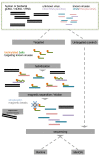
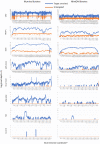
Targeted virome enrichment and sequencing (VirCapSeq-VERT) utilizes a pool of oligos (baits) to enrich all known—up to 2015—vertebrate-infecting viruses, increasing their detection sensitivity. The hybridisation of the baits to the target sequences can be partial, thus enabling the detection and genomic reconstruction of novel pathogens with <40% genetic diversity compared to the strains used for the baits’ design. In this study, we deploy this method in multiplexed mixes of viral extracts, and we assess its performance in the unbiased detection of DNA and RNA viruses after cDNA synthesis. We further assess its efficiency in depleting various background genomic material. Finally, as a proof-of-concept, we explore the potential usage of the method for the characterization of unknown, emerging human viruses, such as SARS-CoV-2, which may not be included in the baits’ panel. We mixed positive samples of equimolar DNA/RNA viral extracts from SARS-CoV-2, coronavirus OC43, cytomegalovirus, influenza A virus H3N2, parvovirus B19, respiratory syncytial virus, adenovirus C and coxsackievirus A16. Targeted virome enrichment was performed on a dsDNA mix, followed by sequencing on the NextSeq500 (Illumina) and the portable MinION sequencer, to evaluate its usability as a point-of-care (PoC) application. Genome mapping assembly was performed using viral reference sequences. The untargeted libraries contained less than 1% of total reads mapped on most viral genomes, while RNA viruses remained undetected. In the targeted libraries, the percentage of viral-mapped reads were substantially increased, allowing full genome assembly in most cases. Targeted virome sequencing can enrich a broad range of viruses, potentially enabling the discovery of emerging viruses.
Keywords: COVID-19; NGS diagnostics; SARS-CoV-2; emerging viruses; nanopore sequencing; target enrichment; virome sequencing.
Conflict of interest statement
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

References
-
- World Health Organization . A Brief Guide to Emerging Infectious Diseases and Zoonoses. WHO Regional Office for South-East Asia; New Delhi, India: 2014. Regional Office for South-East Asia.
-
- Falcinelli S.D., Chertow D.S., Kindrachuk J. Integration of Global Analyses of Host Molecular Responses with Clinical Data to Evaluate Pathogenesis and Advance Therapies for Emerging and Re-Emerging Viral Infections. ACS Infect. Dis. 2016;2:787–799. doi: 10.1021/acsinfecdis.6b00104. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

