Borneol Ester Derivatives as Entry Inhibitors of a Wide Spectrum of SARS-CoV-2 Viruses
- PMID: 35746766
- PMCID: PMC9228966
- DOI: 10.3390/v14061295
Borneol Ester Derivatives as Entry Inhibitors of a Wide Spectrum of SARS-CoV-2 Viruses
Abstract
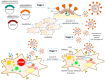
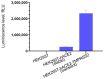
In the present work we studied the antiviral activity of the home library of monoterpenoid derivatives using the pseudoviral systems of our development, which have glycoproteins of the SARS-CoV-2 virus strains Wuhan and Delta on their surface. We found that borneol derivatives with a tertiary nitrogen atom can exhibit activity at the early stages of viral replication. In order to search for potential binding sites of ligands with glycoprotein, we carried out additional biological tests to study the inhibition of the re-receptor-binding domain of protein S. For the compounds that showed activity on the pseudoviral system, a study using three strains of the infectious SARS-CoV-2 virus was carried out. As a result, two leader compounds were found that showed activity on the Wuhan, Delta, and Omicron strains. Based on the biological results, we searched for the potential binding site of the leader compounds using molecular dynamics and molecular docking methods. We suggested that the compounds can bind in conserved regions of the central helices and/or heptad repeats of glycoprotein S of SARS-CoV-2 viruses.
Keywords: SARS-CoV-2; borneol; coronavirus surface protein S-spike; molecular docking; molecular dynamics; pseudoviral system; terpene.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

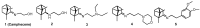

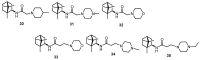

Similar articles
-
(+)-Usnic Acid and Its Derivatives as Inhibitors of a Wide Spectrum of SARS-CoV-2 Viruses.Viruses. 2022 Sep 29;14(10):2154. doi: 10.3390/v14102154. Viruses. 2022. PMID: 36298709 Free PMC article.
-
Simulation of Molecular Dynamics of SARS-CoV-2 S-Protein in the Presence of Multiple Arbidol Molecules: Interactions and Binding Mode Insights.Viruses. 2022 Jan 10;14(1):119. doi: 10.3390/v14010119. Viruses. 2022. PMID: 35062323 Free PMC article.
-
The binding of heparin to spike glycoprotein inhibits SARS-CoV-2 infection by three mechanisms.J Biol Chem. 2022 Feb;298(2):101507. doi: 10.1016/j.jbc.2021.101507. Epub 2021 Dec 18. J Biol Chem. 2022. PMID: 34929169 Free PMC article.
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Structure based Drug Designing Approaches in SARS-CoV-2 Spike Inhibitor Design.Curr Top Med Chem. 2022;22(29):2396-2409. doi: 10.2174/1568026623666221103091658. Curr Top Med Chem. 2022. PMID: 36330617 Review.
Cited by
-
Discovery of N-Containing (-)-Borneol Esters as Respiratory Syncytial Virus Fusion Inhibitors.Pharmaceuticals (Basel). 2022 Nov 11;15(11):1390. doi: 10.3390/ph15111390. Pharmaceuticals (Basel). 2022. PMID: 36422520 Free PMC article.
-
Structural and Functional Characterization of Medicinal Plants as Selective Antibodies towards Therapy of COVID-19 Symptoms.Antibodies (Basel). 2024 May 7;13(2):38. doi: 10.3390/antib13020038. Antibodies (Basel). 2024. PMID: 38804306 Free PMC article.
-
Synthesis and Antiviral Properties of Camphor-Derived Iminothiazolidine-4-Ones and 2,3-Dihydrothiazoles.Molecules. 2022 Jul 25;27(15):4761. doi: 10.3390/molecules27154761. Molecules. 2022. PMID: 35897931 Free PMC article.
-
The Potential of Usnic-Acid-Based Thiazolo-Thiophenes as Inhibitors of the Main Protease of SARS-CoV-2 Viruses.Viruses. 2024 Jan 31;16(2):215. doi: 10.3390/v16020215. Viruses. 2024. PMID: 38399993 Free PMC article.
-
(+)-Usnic Acid and Its Derivatives as Inhibitors of a Wide Spectrum of SARS-CoV-2 Viruses.Viruses. 2022 Sep 29;14(10):2154. doi: 10.3390/v14102154. Viruses. 2022. PMID: 36298709 Free PMC article.
References
-
- WHO . COVID-19 Weekly Epidemiological Update. World Health Organisation; Geneva, Switzerland: 2022. pp. 1–23.
-
- Novikov F.N., Stroylov V.S., Svitanko I.V., Nebolsin V.E. Molecular basis of COVID-19 pathogenesis. Russ. Chem. Rev. 2020;89:858–878. doi: 10.1070/RCR4961. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

