Expression of Alphavirus Nonstructural Protein 2 (nsP2) in Mosquito Cells Inhibits Viral RNA Replication in Both a Protease Activity-Dependent and -Independent Manner
- PMID: 35746799
- PMCID: PMC9228716
- DOI: 10.3390/v14061327
Expression of Alphavirus Nonstructural Protein 2 (nsP2) in Mosquito Cells Inhibits Viral RNA Replication in Both a Protease Activity-Dependent and -Independent Manner
Abstract
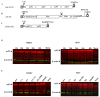
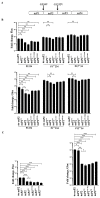
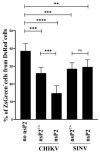
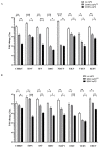
Alphaviruses are positive-strand RNA viruses, mostly being mosquito-transmitted. Cells infected by an alphavirus become resistant to superinfection due to a block that occurs at the level of RNA replication. Alphavirus replication proteins, called nsP1-4, are produced from nonstructural polyprotein precursors, processed by the protease activity of nsP2. Trans-replicase systems and replicon vectors were used to study effects of nsP2 of chikungunya virus and Sindbis virus on alphavirus RNA replication in mosquito cells. Co-expressed wild-type nsP2 reduced RNA replicase activity of homologous virus; this effect was reduced but typically not abolished by mutation in the protease active site of nsP2. Mutations in the replicase polyprotein that blocked its cleavage by nsP2 reduced the negative effect of nsP2 co-expression, confirming that nsP2-mediated inhibition of RNA replicase activity is largely due to nsP2-mediated processing of the nonstructural polyprotein. Co-expression of nsP2 also suppressed the activity of replicases of heterologous alphaviruses. Thus, the presence of nsP2 inhibits formation and activity of alphavirus RNA replicase in protease activity-dependent and -independent manners. This knowledge improves our understanding about mechanisms of superinfection exclusion for alphaviruses and may aid the development of anti-alphavirus approaches.
Keywords: RNA replication; alphaviruses; mosquito; nsP2; protease; superinfection exclusion.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design, execution, interpretation or writing of the study.
Figures
References
-
- Nasar F., Palacios G., Gorchakov R.V., Guzman H., Da Rosa A.P., Savji N., Popov V.L., Sherman M.B., Lipkin W.I., Tesh R.B., et al. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc. Natl. Acad. Sci. USA. 2012;109:14622–14627. doi: 10.1073/pnas.1204787109. - DOI - PMC - PubMed
-
- Vega-Rúa A., Zouache K., Girod R., Failloux A.B., Lourenço-de-Oliveira R. High level of vector competence of Aedes aegypti and Aedes albopictus from ten American countries as a crucial factor in the spread of chikungunya virus. J. Virol. 2014;88:6294–6306. doi: 10.1128/JVI.00370-14. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

