Detection of Ancient Viruses and Long-Term Viral Evolution
- PMID: 35746807
- PMCID: PMC9230872
- DOI: 10.3390/v14061336
Detection of Ancient Viruses and Long-Term Viral Evolution
Abstract
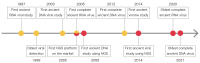
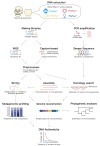
The COVID-19 outbreak has reminded us of the importance of viral evolutionary studies as regards comprehending complex viral evolution and preventing future pandemics. A unique approach to understanding viral evolution is the use of ancient viral genomes. Ancient viruses are detectable in various archaeological remains, including ancient people's skeletons and mummified tissues. Those specimens have preserved ancient viral DNA and RNA, which have been vigorously analyzed in the last few decades thanks to the development of sequencing technologies. Reconstructed ancient pathogenic viral genomes have been utilized to estimate the past pandemics of pathogenic viruses within the ancient human population and long-term evolutionary events. Recent studies revealed the existence of non-pathogenic viral genomes in ancient people's bodies. These ancient non-pathogenic viruses might be informative for inferring their relationships with ancient people's diets and lifestyles. Here, we reviewed the past and ongoing studies on ancient pathogenic and non-pathogenic viruses and the usage of ancient viral genomes to understand their long-term viral evolution.
Keywords: NGS; PCR; TDRP; ancient DNA; ancient RNA; ancient virome; ancient virus; bioinformatics; viral evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

