The Viral Susceptibility of the Haloferax Species
- PMID: 35746816
- PMCID: PMC9229481
- DOI: 10.3390/v14061344
The Viral Susceptibility of the Haloferax Species
Abstract
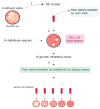
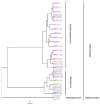
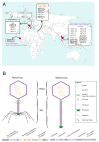
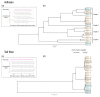
Viruses can infect members of all three domains of life. However, little is known about viruses infecting archaea and the mechanisms that determine their host interactions are poorly understood. Investigations of molecular mechanisms of viral infection rely on genetically accessible virus-host model systems. Euryarchaea belonging to the genus Haloferax are interesting models, as a reliable genetic system and versatile microscopy methods are available. However, only one virus infecting the Haloferax species is currently available. In this study, we tested ~100 haloarchaeal virus isolates for their infectivity on 14 Haloferax strains. From this, we identified 10 virus isolates in total capable of infecting Haloferax strains, which represented myovirus or siphovirus morphotypes. Surprisingly, the only susceptible strain of all 14 tested was Haloferax gibbonsii LR2-5, which serves as an auspicious host for all of these 10 viruses. By applying comparative genomics, we shed light on factors determining the host range of haloarchaeal viruses on Haloferax. We anticipate our study to be a starting point in the study of haloarchaeal virus-host interactions.
Keywords: Haloferax; Haloferax gibbonsii LR2-5; archaeal virus; haloarchaea; host range.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Baquero D.P., Liu Y., Wang F., Egelman E.H., Prangishvili D., Krupovic M. Structure and Assembly of Archaeal Viruses. 1st ed. Volume 108. Elsevier Inc.; Amsterdam, The Netherlands: 2020. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

